Specific and Sensitive Primers Developed by Comparative Genomics to Detect Bacterial Pathogens in Grains
Article information
Abstract
Accurate and rapid detection of bacterial plant pathogen is the first step toward disease management and prevention of pathogen spread. Bacterial plant pathogens Clavibacter michiganensis subsp. nebraskensis (Cmn), Pantoea stewartii subsp. stewartii (Pss), and Rathayibacter tritici (Rt) cause Goss’s bacterial wilt and blight of maize, Stewart’s wilt of maize and spike blight of wheat and barley, respectively. The bacterial diseases are not globally distributed and not present in Korea. This study adopted comparative genomics approach and aimed to develop specific primer pairs to detect these three bacterial pathogens. Genome comparison among target pathogens and their closely related bacterial species generated 15–20 candidate primer pairs per bacterial pathogen. The primer pairs were assessed by a conventional PCR for specificity against 33 species of Clavibacter, Pantoea, Rathayibacter, Pectobacterium, Curtobacterium. The investigation for specificity and sensitivity of the primer pairs allowed final selection of one or two primer pairs per bacterial pathogens. In our assay condition, a detection limit of Pss and Cmn was 2 pg/μl of genomic DNA per PCR reaction, while the detection limit for Rt primers was higher. The selected primers could also detect bacterial cells up to 8.8 × 103 cfu to 7.84 × 104 cfu per gram of grain seeds artificially infected with corresponding bacterial pathogens. The primer pairs and PCR assay developed in this study provide an accurate and rapid detection method for three bacterial pathogens of grains, which can be used to investigate bacteria contamination in grain seeds and to ultimately prevent pathogen dissemination over countries.
The economic impact of plant diseases caused by bacterial pathogens escalates with global warming (Castillo and Plata, 2016) with an increase in international exchange of grains and vegetables among countries (Nutter and Madden, 2009). Comparatively, a number of bacterial pathogens prefer higher temperatures, for infection, than the fungal pathogens. The magnitude of risk for bacterial pathogens dissemination is directly proportional to the massive global exchange of the agricultural products (Pautasso et al., 2010). Therefore, precise measures for plant quarantine become important to restrict dissemination of bacterial plant pathogen globally to ensure secure agricultural production.
Bacterial pathogens, Clavibacter michiganensis subsp. nebraskensis (Cmn), Pantoea stewartii subsp. stewartii (Pss), Rathayibacter tritici (Rt, formerly Clavibacter tritici), are not globally distributed and are thus quarantined pathogens in many countries including Korea. Cmn is a Gram-positive bacterial pathogen causing Goss’s bacterial wilt and blight of maize (Zea mays L.) (Vidaver and Mandel, 1974). The disease shows two different symptoms: a leaf blight and a systematic vascular wilt (Agarkova et al., 2011). Since the first report of this disease in 1969 in the USA, this disease is only limited in USA and Canada (Agarkova et al., 2011; Eggenberger et al., 2016; Vidaver and Mandel, 1974). Transmission of this disease is either through infected seeds or bacteria-infested residues. Rate of transmission via seeds is lower than bacteria-infested residues which seem to be the primary source of inoculum in disease spread (Smidt and Vidaver, 1986). Diagnosis and detection of Goss’s wilt and bacterial pathogen are currently dependent on disease symptoms, immunological assays and selective medium for bacterial pathogen.
Pss causes Stewart’s wilt of maize, mostly transmitted by the corn flea beetle (Chaetoconema pulicaria) and infected seeds (Michener et al., 2002) and the disease is under plant quarantine regulations in more than 60 countries on maize seeds imports from affected areas (Pataky and Ikin, 2003). Symptoms of Stewart’s wilt are somewhat similar to Goss’s wilt by Cmn and thus a careful examination is necessary to distinguish the two diseases. To differentiate plant-associated Pantoea species, several methods have been developed. Immunoassay or polymerase chain reaction (PCR) primers have been described (Coplin et al., 2002; Lamka et al., 1991; Tambong et al., 2008; Wensing et al., 2010). However, those analyses cannot completely distinguish species of Pantoea or subspecies of P. stewartii (Wensing et al., 2014). Recently, Wensing et al. (2014) developed a single-nucleotide polymorphism based PCR assay to differentiate Pss from P. stewartii subsp. indologenes.
Rt is a Gram-positive coryneform bacterial pathogen causing spike blight, also called yellow ear rot or Tundu disease, in wheat and barley (Paruthi and Gupta, 1987). Rt infection and rapid spread is frequently associated with Anguina tritici, a nematode, producing seed galls (ear cockles) in certain plant varieties (Paruthi and Bhatti, 1985). Despite the wide distribution of this bacterial pathogen (Park et al., 2017), no reliable detection method has been reported so far.
DNA-based diagnostic methods such as PCR have been widely used as an effective approach to detect bacterial pathogens from diseased plants, which offer advantages over traditional antibody-based detections which are expensive and time consuming processes (Cho et al., 2011; De Boer et al., 1988; Gudmestad et al., 2009; Vincelli and Tisserat, 2008). Either the 16S rRNA gene or the spacer region between the 16S and 23S rRNA genes of bacterial pathogen, known phylogenetic markers, have been primarily targeted for amplification by PCR (Kang et al., 2016). The possibility for use of these sequences to detect bacterial pathogens remains technically limited to sensitively distinguish bacteria at species or below species level (Baker et al., 2003; Henriques et al., 2012). Hence, there are growing efforts to search for new marker gene or sequences to be developed as detection markers for bacterial pathogens of plants with high reliability. A perpetual increase in the availability of whole genome sequences of bacterial pathogens has shifted the attention to use of comparative genome analysis, which is the most effective ways to select candidate marker gene of bacterial pathogens (Ahmed et al., 2008; Demkin et al., 2017; Papaiakovou et al., 2017; Tao et al., 2015).
Since there are no primers available to detect bacterial pathogens, Cmn and Rt, we aimed to develop specific and sensitive primers to detect bacterial pathogens in grains. In this study, by taking advantage of comparative genome analysis of bacterial pathogens, we assessed several candidate primers to detect three bacterial pathogens of grains, and selected the specific and sensitive primer pairs for these bacterial pathogens.
Materials and Methods
Bacterial strains and culture condition
Most of bacterial strains used in this study were obtained from the National Collection of Plant Pathogenic Bacteria (NCPPB, York, UK) and the Korean Agricultural Culture Collection (KACC, Korea) (Table 1). Bacterial strains of C. michiganensis subsp. capsici (Cmc) (Oh et al., 2016) and Cmm (Yim et al., 2012) were kindly provided from Chang-Sik Oh at the Kyung Hee University, and Jae-Soon Cha at the Chungbuk National University, Korea, respectively. All of bacterial strains from NCPPB are listed as plant quarantine bacteria in Korea and many other countries, therefore, these strains were obtained under official permission from Animal and Plant Quarantine Agency, Korea for use only in a restricted area during limited time period.
Both Clavibacter species and Pectobacterium species were grown at 25°C while Pantoea species were grown at 30°C in nutrient broth (NB). The Rathayibacter species were grown in nutrient broth yeast extract broth (NBY) (Hayward, 1964) at 25°C. Curtobacterium flaccumfaciens pv. flaccumfaciens (Cff ) was grown in yeast peptone glucose agar medium (Vidaver, 1967) at 30°C. Genomic DNA of bacterial strains was purified using a Wizard Genomic DNA Purification kit (Promega, Fitchburg, WI, USA) and the concentration of purified DNA was determined by using a NanoDrop2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
Primer design by comparative genomic analysis
To search for pathogen-specific genes of target bacterial pathogens, whole genome information of target bacteria and their closely related species was retrieved from National Center for Biotechnology Information (NCBI) database (ftp://ftp.ncbi.nlm.nih.gov/genomes/bacteria/) at completed or scaffold level of assembly except for contig level and is listed in Table 2. We also used whole genome sequence information of Rt previously studied (Park et al., 2017). Based on the acquired genomic information, BLASTN searches were initially performed using 2,801 coding sequences (CDS) of Cmn, 4,942 CDS of Pss and 2,995 CDS of Rt as query against the constructed subject nucleotide sequences consisting of CDS of closely related species to the target bacterial pathogen. To eliminate genes highly matched to non-target bacteria, CDS with the lowest e-value < 0.1 were discarded, the filtered CDS were reran and searched against the NCBI’s non-redundant (nr) DNA sequence database (Yu et al., 2010). The genes including non-matched CDS with e-value ≥ 0.05, query coverage and identity < 30% were selected as candidates of the target bacterial pathogen-specific genes.
To find a region to be amplified by PCR experiments, the specific genes more than 500 bp were compared individually to the nr database in the NCBI BLAST web site (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Subsequently, primers were designed from the unmatched region using Primer3 program (Rozen and Skaletsky, 2000). Finally, specificity of the designed candidate primer pairs were predicted in silico by searching against the nr sequences via Primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast) of NCBI with default option.
Polymerase chain reaction (PCR) and quantitative PCR (qPCR)
Primer pairs used for PCR and qPCR amplification reactions are listed in Supplementary Table 1. Thermal cycling for PCR was performed according to the following program: an initial denaturation for 5 min at 95°C, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at the specified temperature (Supplementary Table 1) for 30 s, and a final extension at 72°C for 1 min. PCR reaction mixture included all required components in a 50 μl of reaction volume with 150 ng of template genomic DNA. Negative control included PCR reaction without template DNA. When necessary for PCR sensitivity assay, template genomic DNA was serially diluted from 150 ng to 0.1 ng.
qPCR was performed with reaction mixture containing SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) and various concentrations of template genomic DNA at the following program: an initial denaturation for 2 min at 95°C, followed by 39 cycles of denaturation at 95°C for 5 s, annealing at the specified temperature (Supplementary Table 1) for 20 s, and a final extension at 72°C for 30 min. The qPCR data were displayed using the CFX Manager Software ver. 3.1. Each reaction was performed in triplicate. qPCR result of individual gene was evaluated using the iCycler iQ Real-Time PCR Detection System. The Ct values of qPCR products of each gene were used to generate a standard curve against the number of gene copy (equivalent to the number of bacteria) used for qPCR (Supplementary Fig. 1–3). The number of gene (or genome) copy was obtained by dividing the amount of used DNA for qPCR (weight) with a molar weight of genomic DNA (genome size (bp) × 660 g/bp), and multiplying by 6.02 × 1023 molecules.
PCR detection of bacteria on seeds
To detect bacterial pathogen from seeds, we followed a previously described method with slight modification (An et al., 2015). Clean corn seeds and dehulled wheat seeds were purchased and used to detect bacterial pathogens in the seeds by PCR. The seeds were surface sterilized by immersing in 70% ethanol for 30 min and subsequently in 1% sodium hypochlorite solution for 10 min. Finally, the sterilized seeds were extensively washed 5 times with sterile distilled water, completely dried under a clean bench, and stored at 4°C until use. To artificially inoculate bacterial pathogens Cmn, Pss, Rt, 20 gram of surface sterilized corn seeds and 10 gram of wheat seeds were incubated by shaking at 100 rpm in a bacterial culture suspension (3 fold volume of seed volume with 108 cfu/ml) for 1 h. The seeds were harvested by removing the bacterial suspension and dried under clean bench prior to storage at 4°C. The surface sterilized seeds were used as negative controls.
To detect bacterial pathogens from the seeds, 10 gram of seeds was shaken at 180 rpm for 5 min in sterile distilled water containing a drop of Tween 20. The suspension was collected to determine the number of inoculated bacteria per seeds. Finally, the suspension was boiled at 95°C for 10 min, prior to PCR amplification to detect the presence of bacterial pathogen in the seeds.
Results and Discussion
Candidate primers generated by comparative genomics and specific detection of bacterial pathogens
Using comparative genomics among phylogenetically related bacterial pathogens, Cmn, Pss and Rt, we generated 15–20 candidate primer pairs per bacterial pathogen to detect the corresponding bacterial pathogens (data not shown). Our first round of screening yielded the specific amplification of expected PCR products from the target bacterial pathogens (data not shown). All of the primer pairs which showed specific detection among the tested primers are listed in Table S1. Based on specificity and sensitivity, we further selected 2 primer pairs for each bacterial pathogen, namely: Cmn-3 (Fig. 1A) and Cmn-4 (Fig. 1B) for Cmn; Pss2-4 (Fig. 2A) and Pss2-9 (Fig. 2B) for Pss; Rt-4 (Fig. 3A) and Rt-5 (Fig. 3B) for Rt.
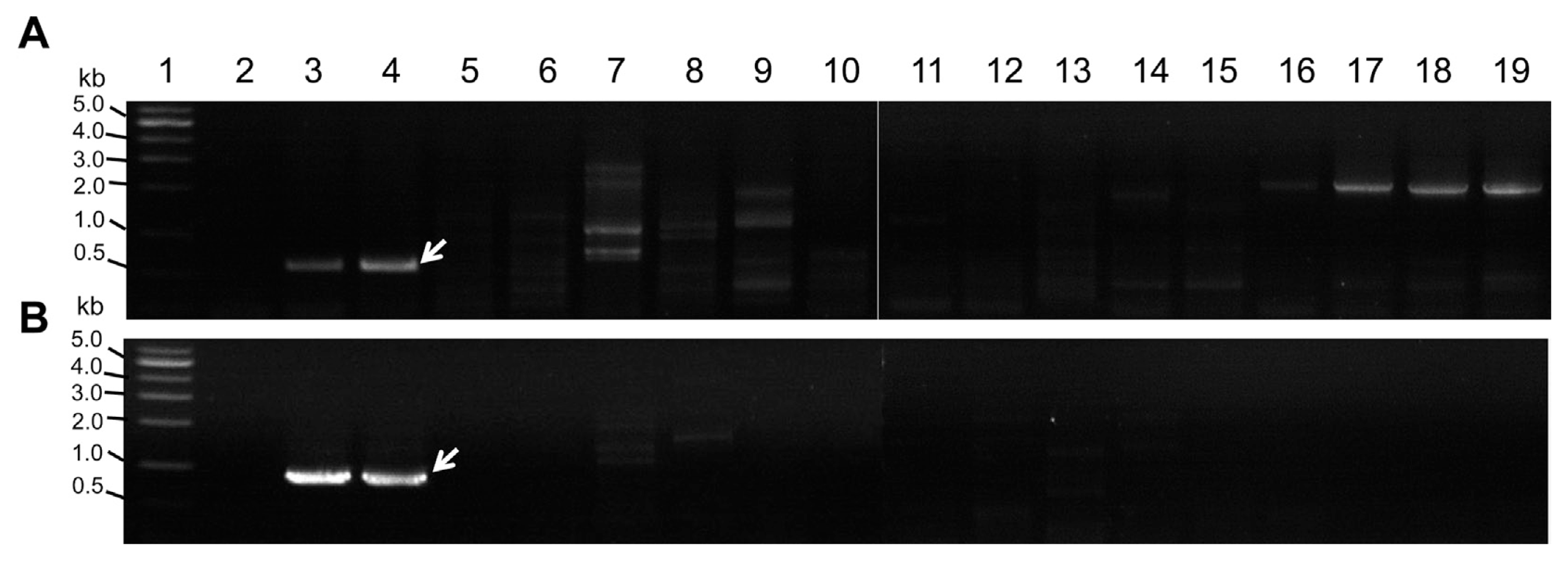
Amplification of expected PCR product from Clavibacter michiganensis subsp. nebraskensis (Cmn) using two primer pairs. (A) Amplification by Cmn-3, (B) amplification by Cmn-4. Lane 1, DNA size standard (1 kb ladder); lane 2, no template DNA; lane 3, Cmn NCPPB 2581; lane 4, Cmn NCPPB 2582; lane 5, Rathayibacter tritici (Rt) NCPPB 255; lane 6, Rt NCPPB 471; lane 7, R. toxicus NCPPB 3552; lane 8, R. rathayi NCPPB 2980; lane 9, R. iranicus NCPPB 2253; lane10, C. michiganensis subsp. insidiosus NCPPB 1109; lane 11, C. michiganensis subsp. tesselarius NCPPB 3664; lane 12, Rt NCPPB 1857; lane 13, Rt NCPPB 1953; lane 14, C. michiganensis subsp. michiganensis (Cmm) NCPPB LMG7333; lane 15, Cmm PPP1; lane 16, C. michiganensis subsp. capsici (Cmc) PF003; lane 17, Cmc PF005; lane 18, Cmc PS006; lane 19, Cmc PF008. The white arrow represents the expected PCR product.

Specific detection of Pantoea stewartii subsp. stewartii (Pss) using two primer pairs. (A) Amplification by Pss2-4, (B) amplification by Pss2-9. Lane 1, DNA size standard (1 kb ladder); lane 2, no template DNA; lane3, P. stewartii subsp. indologens (Psi) NCPPB 2280; lane 4, Pss NCPPG 1553; lane 5, Pss NCPPB 3253; lane 6, P. ananatis pv. ananatis (Paa) NCPPB 3619; lane 7, Psi NCPPB 228; lane 8, P. agglomerans (Pa) NCPPB 10054; lane 9, Pa NCPPB 15275; lane 10, Pectobacterium carotovorum subsp. carotovorum (Pcc) KACC 10057; lane 11, Pcc KACC 10225; lane 12, Pcc KACC 10408; lane 13, Pcc KACC 10421; lane 14, Pcc KACC 10458; lane 15, Pss NCPPB 2295; lane 16, Pss NCPPB 3379. The arrow on the right represents the expected PCR product.

Specific amplification of PCR product from Rathayibacter tritici (Rt) using selected two primer pairs. (A) Amplification by Rt-4, (B) amplification by Rt-5. Lane 1, DNA size standard (1 kb ladder); lane 2, no template DNA; lane 3, Rt NCPPB 1857; lane 4, Rt NCPPB 1953; lane 5, Rt NCPPB 255; lane 6, Rt NCPPB 471; lane 7, C. michiganensis subsp. nebraskensis (Cmn) NCPPB 2581; lane 8, Cmn NCPPB 2582; lane 9, R. toxicus NCPPB 3552; lane 10, R. rathayi NCPPB 2980; lane 11, R. iranicus NCPPB 2253; lane 12, C. michiganensis subsp. insidiosus NCPPB 1109; lane 13, C. michiganensis subsp. tesselarius NCPPB 3664; lane 14, C. michiganensis subsp. michiganensis (Cmm) LMG 7333; lane 15, Cmm PPP1; lane 16, C. michiganensis subsp. capsici (Cmc) PF003; lane 17, Cmc PF005; lane 18, Cmc PS006; lane 19, Cmc PF008. The white arrow represents the expected PCR product.
Since Rt was previously classified as a species of Clavibacter and separated from Clavibacter (Zgurskaya et al., 1993), we tested the primer pairs with Clavibacter species. Most of primer pairs designed by Rt genome information (Park et al., 2017) seem specific and neither detect the Clavibacter species nor the closely related Rathayibacter species such as R. toxicus, R. iranicus and R. rathayi (Fig. 3). Pss and many Pantoea species previously belonged to Erwinia species and were closely related to Erwinia species (Mergaert et al., 1993), which include Pectobacterium carotovorum (previously E. carotovora). Therefore, we tested Pss primer pairs with Pantoea species and P. carotovorum strains. PCR with Pss2-4 and Pss2-9 primer pairs resulted in the specific amplification from Pss strains which differed from other Pantoea species and P. carotovorum strains (Fig. 2).
Most of the primer pairs targeted amplification of genes encoding hypothetical proteins except Pss2-4, which amplified the gene encoding antiterminator Q of prophage of Pss. In case of the gene for antiterminator Q, it could be strain-specific and may not be considered suitable for specific detection. However, all of the 4 strains tested for Pss exhibited the specific amplification of the gene (Fig. 2A), suggesting that the gene encoding antiterminator Q would be a good marker for Pss detection. Other primer pairs which targeted the gene encoding hypothetical proteins should be carefully evaluated with more target bacterial strains. Genes encoding bacterial virulence determinants, such as toxins and plant cell wall degrading enzymes (CWDE), could serve as bacteria specific targets for detection. However, our preliminary comparative genome analysis indicated that genes for the virulence factors are not bacterial species or subspecies-specific, which can mislead for detection of bacterial pathogens from grains. In fact, it was previously reported that several bacterial genes encoding toxins or CWDE are conserved among different bacterial species (Lee et al., 2016; Murray et al., 2017).
Sensitivity of the selected primers
While most of candidate primer pairs amplified the expected PCR products from target pathogens but not from other related bacterial strains, the sensitivity seemed to be variable from each primer pair against the target pathogens. Therefore, we tested sensitivity of the primer pairs using serially diluted templated DNA from the type strain of three bacterial pathogens Cmn, Pss, Rt, and an additional strain in the same species for Cmn and Rt. At the indicated PCR condition, primers Cmn-3 and Cmn-4 to detect Cmn (Fig. 4A), Pss2-4 and Pss2-9 to detect Pss (Fig. 4B), were highly sensitive to show apparent amplification at 0.1 ng of templated DNA (2 × 10−3 ng/μl). However, primer pairs Rt-4 and Rt-5 showed less sensitivity to detect Rt at the equivalent amount of template DNA (Fig. 4C). Sensitivity of the primer pairs was further investigated by qPCR with ten-fold diluted DNA of target bacteria. The tested two primer pairs for each bacterium showed similar sensitivity against target bacteria (data not shown).

Primer sensitivity assay using serially diluted genomic DNA of plant pathogenic bacteria. (A) Sensitive detection of Clavibacter michiganensis subsp. nebraskensis (Cmn), (B) sensitive detection of Pantoea stewartii subsp. stewartii (Pss), (C) sensitive detection of Rathayibacter tritici (Rt). Primer pairs and used bacterial strains of were indicated. Lane 1: DNA size standard; lane 2, no genomic DNA; lane 3, genomic DNA 150 ng; lane 4, genomic DNA 100 ng; lane 5, genomic DNA 50 ng; lane 6, genomic DNA 10 ng; lane 7, genomic DNA 1 ng; lane 8, genomic DNA 0.1 ng.
Overall, our specificity and sensitivity assay with candidate primer pairs generated from comparative genome analysis revealed that most of primers were specific to amplify the target gene from the corresponding pathogenic bacteria, suggesting the gene probing from genome-based analysis is a promising tool to provide pathogen specific markers. However, the sensitivity of primers to detect target pathogen varied, which will need additional work to select best primer pairs. It will be more complicated if we consider plant component and plant genome to detect bacterial pathogen resided inside of plant seeds.
Detection of bacterial pathogens in artificially inoculated seeds
To test if the primer pairs developed in this study could be used to detect the target pathogen from seeds of corn and wheat effectively, we used seeds artificially inoculated with target bacteria. This is because all the three pathogens have not been reported in Korea, and seeds infected with Cmn, Pss, and Rt were not available. Among the primer pairs tested, three primer pairs, Rt-4, Cmn-3 and Pss2-4, exhibited the specific detection of bacteria from the artificially inoculated seeds (Fig. 5). The number of target bacteria on the corn seeds was enumerated for Cmn as 8.8 × 103 cfu/g seed, for Pss as 7.7 × 104 cfu/g seed, and for Rt on wheat seeds as 7.84 × 104 cfu/g seed. These results indicated that the selected primer pairs can detect target bacterial pathogens that are inhabited on the surface of seeds at the level of 103 to 104 cells per gram of seeds. No amplification of specific bacteria was detected from the sterilized seeds without bacterial inoculation or seeds not surface sterilized (Fig. 5). The seeds were randomly purchased from market and it turned out they might be target pathogen free.

Specific detection of bacteria from the artificially inoculated seeds. (A) Detection of Rathayibacter tritici by Rt-4 on wheat seeds, (B) detection of Clavibacter michiganensis subsp. nebraskensis by Cmn-3 on corn seeds, (C) detection of Pantoea stewartii subsp. stewartii by Pss2-3 on corn seeds. Lane M, DNA size standard (1 kb ladder); S, seeds original; Nt, surface sterilized seeds; Sb, seeds infected with bacteria.
Although we inoculated seeds of corn or wheat with either Cmn, Pss, or Rt, this is not natural assay because when bacteria infest the seeds in natural disease occurrence, the seeds may be both internally and externally contaminated (Rane and Latin 1992; Tancos et al., 2013). This assay was primarily performed using bacterial cells harvested from the surface but not from the total DNA isolated from grinded seeds. If infested seeds harbour pathogenic bacteria mostly inside the seeds, we require total DNA from the grinded seeds as a template, which will interfere with the specificity and sensitivity of primers. This should further be extensively tested with the naturally infected seeds, which was not possible to do in this study because of strict regulations to handle target bacterial pathogen in Korea.
Supplementary data
Acknowledgments
This research was carried out with support by a grant from the Next Generation BioGreen21 (Project No. PJ01313101). Rural Development Administration, and by a grant from the Research of Animal and Plant Quarantine Agency, South Korea.