Phylogenetic Characterization of Tomato chlorosis virus Population in Korea: Evidence of Reassortment between Isolates from Different Origins
Article information
Abstract
Tomato chlorosis virus (ToCV) is a whitefly-transmitted and phloem-limited crinivirus. In 2013, severe interveinal chlorosis and bronzing on tomato leaves, known symptoms of ToCV infection, were observed in greenhouses in Korea. To identify ToCV infection in symptomatic tomato plants, RT-PCR with ToCV-specific primers was performed on leaf samples collected from 11 tomato cultivating areas where ToCV-like symptoms were observed in 2013 and 2014. About half of samples (45.18%) were confirmed as ToCV-infected, and the complete genome of 10 different isolates were characterized. This is the first report of ToCV occurring in Korea. The phylogenetic relationship and genetic variation among ToCV isolates from Korea and other countries were also analysed. When RNA1 and RNA2 are analysed separately, ToCV isolates were clustered into three groups in phylogenetic trees, and ToCV Korean isolates were confirmed to belong to two groups, which were geographically separated. These results suggested that Korean ToCV isolates originated from two independent origins. However, the RNA1 and RNA2 sequences of the Yeonggwang isolate were confirmed to belong to different groups, which indicated that ToCV RNA1 and RNA2 originated from two different origins and were reassorted in Yeonggwang, which is the intermediate point of two geographically separated groups.
Introduction
Tomato chlorosis disease is one of the most devastating diseases in tomato crop production (Hanssen et al., 2010). The chlorotic leaf symptom of tomatoes was observed in Florida in 1989, and the virus responsible was first named Tomato chlorosis virus (ToCV) in the USA (Wisler et al., 1998b). Subsequently, this virus has been distributed to many parts of the world including European, American, African and Asian countries (Abou-Jawdah et al., 2006; Accotto et al., 2001; Alvarez-Ruiz et al., 2007; Arruabarrena et al., 2015; Barbosa et al., 2008; Bese et al., 2011; Castro et al., 2009; Çevik and Erkıß, 2008; Dalmon et al., 2005; Dovas et al., 2002; Fiallo-Olivé et al., 2011; Hirota et al., 2010; Jacquemond et al., 2009; Lett et al., 2009; Louro et al., 2000; Segev et al., 2004; Sundaraj et al., 2011; Wintermantel et al., 2001; Wintermantel and Wisler, 2006; Zhao et al., 2013a). In northeast Asia, ToCV occurrence was originally reported in China, Taiwan and Japan (Hirota et al., 2010; Tsai et al., 2004; Zhao et al., 2013b), but was not reported in Korea until 2013. ToCV is a species of the genus Crinivirus that belongs to the family Closteroviridae with flexuous filamentous particles of approximately 800 to 850 nm in length (Liu et al., 2000; Wisler et al., 1998b). ToCV is usually phloem-limited and is transmitted by whiteflies (Bemisia and Trialeuroides spp.) in a semi-persistent manner (Karasev, 2000; Wisler et al., 1998a). Mechanical inoculation and seed transmission have not been demonstrated. The genome consist of two segments of linear, positive-sense and single-stranded RNA, which are separately encapsidated (Wisler et al., 1998b; Wintermantel et al., 2005; Zhao et al., 2014). The size of ToCV RNAs 1 and 2 is 8595nt and 8247nt, respectively. RNA1 contains four open reading frames (ORFs), which encode proteins for replication. RNA2 codes nine ORFs that express proteins involved in viral encapsidation, movement and vector transmission (Wintermantel et al., 2005).
In tomato, ToCV causes interveinal yellowing that can be observed first on lower leaves and subsequent development of leaf thickening, bronzing and necrotic flecks on the older leaves (Wisler et al., 1998b; Wintermantel et al., 2005). Theses typical patterns gradually proceed toward the growing point. Although no obvious fruit symptoms have been observed, crop yield can be significantly reduced due to the loss of photosynthetic area (Wintermantel et al., 2005). Symptoms caused by ToCV are easily confused with those caused by physiological or nutritional deficiency and are very similar to those of other whitefly-transmitted viruses such as Tomato infectious chlorosis virus (TICV) (Wisler et al., 1998b). So, infection of tomato plants with these viruses is difficult to diagnose based on symptoms. In addition to tomatoes, potato (Solanum tuberosum), sweet pepper (Capsicum annuum) and zinnia (Zinnia elegans) are known ToCV hosts (Barbosa et al., 2010; Fortes and Navas-Castillo, 2012; Lozano et al., 2004; Tsai et al., 2004; Vargas et al., 2011). In total, about 36 species of plants have also been reported as ToCV hosts (Alvarez-Ruiz et al., 2007; Barbosa et al., 2010; Font et al., 2004; Fortes and Navas-Castillo, 2012; Lozano et al., 2004; Morris et al., 2006; Segev et al., 2004; Solórzano-Morales et al., 2011; Trenado et al., 2007; Tsai et al., 2004; Wintermantel and Wisler, 2006; Vargas et al., 2011).
Genetic exchanges through reassortment and recombination are major evolutionary factors for RNA plant viruses (Aranda et al., 1997; Domingo and Holland, 1994; Nagy, 2008; Simon and Bujarski, 1994), that can result in differences in symptom severity, host range, or transmission efficiency of plant viruses (Thekke-Veetil et al., 2015). Virus reassortment is a process of genetic recombination for multipartite (segmented) RNA viruses that occurs in host cells co-infected with multiple viruses and generates hybrid progeny viruses with novel genome combinations (Marshall et al., 2013; Vijaykrishna et al., 2015). Despite the importance of reassortment for plant viruses that can affect the production of economically important crops, many related studies on virus reassortment have focused on influenza virus and other viruses infecting humans or animals (Barton et al., 2014; Fuller et al., 2013; Savory et al., 2014; Wille et al., 2011).
In this study, we report the viral genome sequences of 10 ToCV isolates obtained from Korea, and present results of phylogenetic analyses among these isolates and ToCV isolates from other countries that provide evidence of reassortment of two viral segments originating from two geographically separated groups.
Materials and Methods
Sample collection, total RNA extraction and RT-PCR
In 2013 and 2014, a total of 394 samples of tomato leaves showing interveinal chlorosis and bronzing were collected from 11 tomato cultivation areas (Yeoju, Gwangju, Nonsan, Iksan, Yeonggwang, Hwasun, Hampyeong, Jeju, Seogwipo, Pyeongtaek and Buyeo) (Fig. 1, Table 1). ToCV-specific primers were designed based on the viral genome sequence of isolate Gr-535 RNA2 (EU284744.1) retrieved from the GenBank database using the Primer3 program (Rozen and Skaletsky, 2000). The primer sequences were as follows: ToCV-RNA2-1F (5′-ACCTTGGCAGGTTGTGAAAC-3′) and ToCV-RNA2-1R (5′-CGATATCTGGTGGGAGGCTA-3′). Total RNA was extracted from leaf samples using Viral Gene-spin™ Viral DNA/RNA Extraction Kit (iNtRON Biotechnology, Seongnam, Korea). cDNA was synthesised from extracted total RNA using AMV Reverse Transcriptase (Promega, Madison, WI, USA) and ToCV specific primer (ToCVRNA2-1R). PCR was conducted using a ToCV-specific primer set (ToCV-RNA2-1F and ToCV-RNA2-1R) and GoTaq® Flexi DNA Polymerase (Promega) in a T100™ thermal cycler (Bio-Rad, Hercules, CA, USA) under the following conditions: initial denaturation at 95°C for 3 min; 35 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C; and a final extension step at 72°C for 5 min. The expected size of the RT-PCR product is 827 bp.
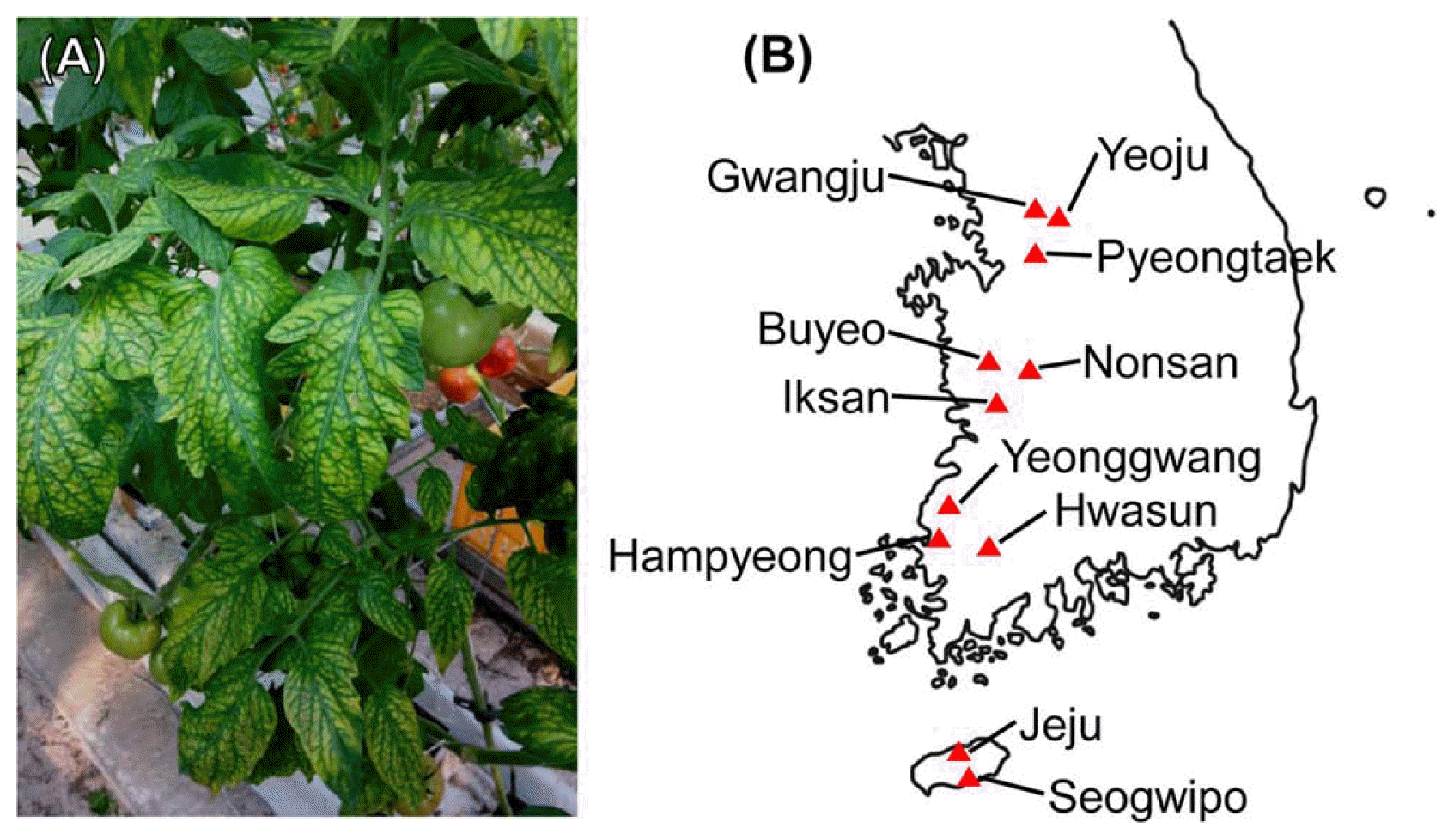
Occurrence of ToCV in Korea. (A) Symptomatic tomato leaves from Iksan showing interveinal leaf chlorosis. (B) Geographic locations of sampling sites.
Full-length genome sequencing of the ToCV Korean isolates
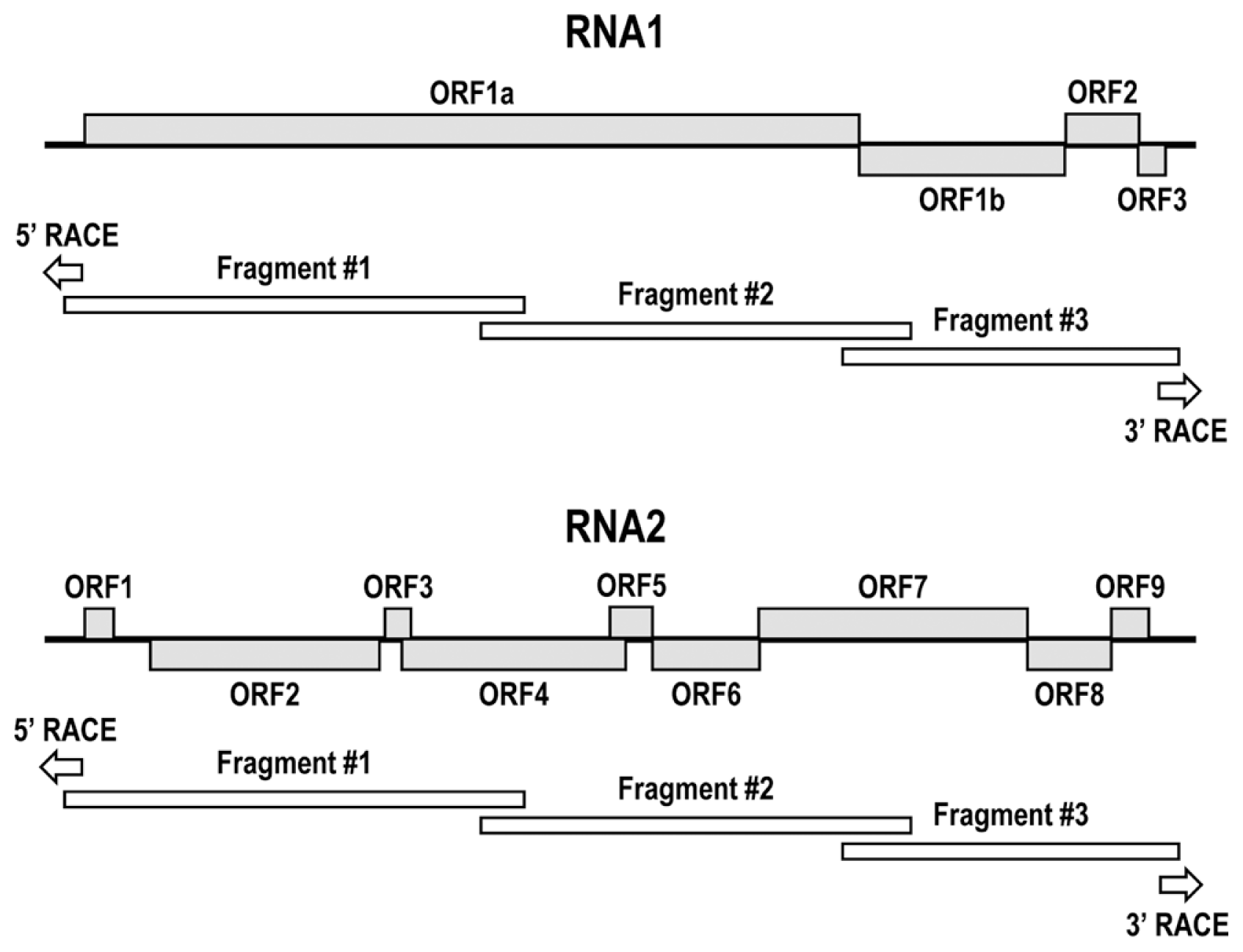
To determine full-length nucleotide sequences, RT-PCR was performed with primers designed based on the previously reported ToCV sequences (GenBank accession numbers: AY903447.1 and KJ815045.1) (Table 2). Viral genome amplification was conducted using LA Taq DNA polymerase (Takara, Tokyo, Japan) in a T100™ thermal cycler (Bio-Rad) under the following conditions: initial denaturation at 95°C for 3 min; 35 cycles of 30 s at 94°C, 30 s at 55°C and 3 min at 72°C; and a final extension step at 72°C for 5 min. Full-length nucleotide sequences of RNA1 and RNA2 were acquired by combining three overlapping RT-PCR products (Fig. 2). Rapid amplification of cDNA ends (RACE) was performed to determine the 5′ and 3′ ends of the viral genomic segments. The amplified PCR products were purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced (Macrogen, Seoul, Korea).
Sequence analysis
Full genome sequences were assembled using the DNASTAR software (DNASTAR, Madison, WI, USA) and compared by a BLAST search (https://blast.ncbi.nlm.nih.gov) with previously reported sequences in the GenBank database (Johnson et al., 2008). Identification of open reading frames was performed by the ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder).
Complete sequences of the 10 ToCV isolates from Korea and eight previously reported isolates was used to examine population genetics. The nucleotide sequences were aligned using the Clustal X and DNAstar programs (Thompson et al., 2002). Phylogenetic analyses of the complete genome sequences were performed by the maximum likelihood method implemented in the MEGA6 program (Tamura et al., 2013). Statistical significance of tree branching was assessed by performing 1000 bootstrap replications. The Geneious software (Biomatters, Auckland, New Zealand) was used to analyse nucleotide identities (Kearse et al., 2012). Pairwise genetic distances was analysed by Kimura’s two-parameter method using the MEGA6 program (Kimura, 1980; Tamura et al., 2013). Database accession numbers and the complete sequences for the isolates used in the phylogenetic and similar analyses in this study are shown in Table 3.
Results and Discussion
Outbreak of viral infection in tomato plants in Korea
In January 2013, tomato plants showing virus-like symptoms of yellowing, bronzing, and chlorosis on lower leaves was found in Iksan, Korea (Fig. 1). Total RNAs were then extracted from all samples and tested by RT-PCR using ToCV specific primers. ToCV-specific amplicons were detected from symptomatic samples in Iksan. To examine distribution and occurrence pattern of ToCV, during the period from 2013 to 2014, a total of 394 samples of symptomatic tomatoes were collected 11 tomato cultivation regions in Korea. Among the 394 samples, 178 were found to be infected with ToCV based on RT-PCR analysis (Table 1). RT-PCR products were sequenced, and BLAST results showed a high sequence identity (> 97%) with previously reported ToCV isolates. Most samples (79.2%) infected with ToCV were co-infected with TYLCV, which showed 99–100% similarity to previously reported sequence of a Korean isolate (GenBank accession number: JN680149.1).
Molecular characterization of Korean ToCV isolates
To examine molecular genetic structure of ToCV population in Korea, the complete nucleotide sequences of 10 ToCV Korean isolates identified in 2013 (except for isolates from Yeoju and Gwangju areas where only one individual was identified) were determined and deposited to the GenBank database (GenBank accession numbers are provided in Table 3). Comparison of the complete nucleotide sequences of the ToCV Korean isolates with all ToCV isolates available on the NCBI database showed overall sequence identities ranging from 97.4 to 99.7% for RNA1 and from 97.5 to 99.7% for RNA2 (data not shown). ToCV RNA1 consisted of four open reading frames (ORFs), as previously reported, and the complete genomic sequences of RNA2 of the ToCV Korean isolates were confirmed as containing nine ORFs (Fig. 2).
Genetic structure of the ToCV population
The MEGA 6.0 program was used to construct a phylogenetic tree using 10 complete sequences determined in this study and eight complete sequences retrieved from the GenBank database (Table 2). Phylogenetic trees constructed using the full-length nucleotide sequences of ToCV RNA1 and RNA2 revealed that the ToCV isolates could be clustered into three groups (Fig. 3). Our results suggested that the ToCV isolates from Jeju, Hwasun, and Hampyeong belong to the same group as isolates from Greece and Brazil (Clade 1), whereas the isolates from Iksan and Nonsan were similar to the isolates from the USA and China (Clade 2) (Fig. 3). In particular, for the isolates from Yeonggwang (ToCV-YG, JN1 and JN2), RNA1 was closer to the isolates from the USA and China in Clade 2 than those from Greece and Brazil, whereas RNA2 showed the opposite tendency, with these isolates in Clade 1 (Fig. 3, 4). From a geographical point of view, the isolates collected from northern areas of Yeonggwang belonged to group 1, while those from the southern area of Iksan were included in group 2 (Fig. 4). This clustering of the ToCV population was further supported by nucleotide diversity analyses. The genetic diversities within and between sub-populations, which were designated based on the phylogenetic trees, were estimated by Kimura’s two-parameter method (Kimura, 1980). The analyses showed that the genetic diversities within subpopulations were somewhat lower compared with those between subpopulations (Table 4). Although ToCV is an RNA virus, population analyses of the ToCV isolates showed high conservation and low molecular variation among the isolates. However, the concatenated sequences of entire genomes (i.e., RNA1 + RNA2) of ToCV isolates strictly indicate that reassortment (Table 4, Fig. 3). Genetic reassortment is an important evolutionary event in the diversification of RNA viruses.
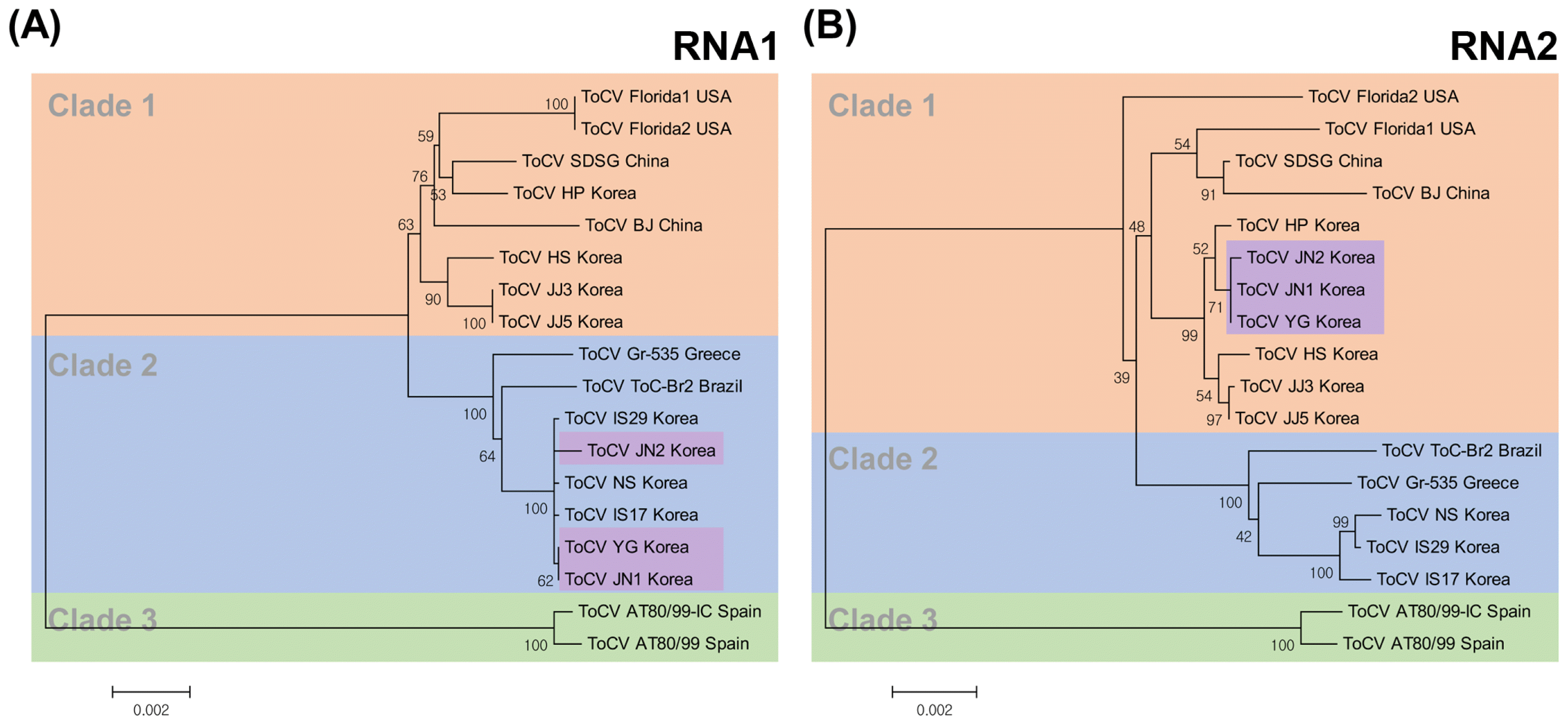
Molecular Phylogenetic analysis of ToCV RNA1 (A) and RNA2 (B) using the maximum likelihood method. The evolutionary history was inferred by using the maximum likelihood method based on the Tamura-Nei model (Tamura and Nei, 1993). The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 18 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013). ToCV isolated were grouped into three clades [clade 1 (red), 2 (blue) and 3 (green)] and two segments (RNA1 and RNA2) belonged to different clades in the case of isolates JN1, JN2 and YG (purple).

Geographic distribution of ToCV isolates in Korea. ToCV isolates from tomato were clustered into three subpopulations based on the phylogenetic analysis.

Genetic diversity of population of ToCV RNA1 and RNA2. Analyses were conducted using the Kimura 2-parameter model (Kimura, 1980). The analysis involved three clades. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013)
In this study, it is confirmed that ToCV isolates in Korea are grouped into two clades based on the phylogenetic analyses. This clustering makes it possible to hypothesize that the ToCV isolates found in Korea have at least two different origins, which can be separated geographically. However, this hypothesis cannot be proven only by the clues provided in this study. In order to prove this, it is necessary to obtain information on the inflow of viruliferous whitefly or ToCV infected plants at the early stage of virus occurrence, but it is not easy to confirm this.
We also found that genetic exchanges have occurred by segment reassortment in natural ToCV populations. Phylogenetic analyses of three Korean isolates (ToCV YG, JN1 and JN2) provided a significant clue to reassortment between two different groups. This means that another ressortment inducing more severe economic damage may occur when ToCV strain(s) that differ from those previously reported arise and are introduced.
Acknowledgments
This research was supported by a grant from the Agenda Program (PJ012013) funded by the Rural Development Administration of Korea and a fund (Project Code No. Z-1543086-2017-21-01) by Research of Animal and Plant Quarantine Agency, South Korea.