 |
 |
| Plant Pathol J > Volume 34(5); 2018 > Article |
|
Abstract
The red flour beetle, Tribolium castaneum, is one of the most common and economically important pests of stored cereal products worldwide. Furthermore, these beetles can act as vectors for several fungal post-harvest diseases. In this study, we collected T. castaneum from 49 rice processing complexes (RPCs) nationwide during 2016-2017 and identified contaminating fungal species on the surface of the beetles. Five beetles from each region were placed on potato dextrose agar media or Fusarium selection media after wet processing with 100% relative humidity at 27┬░C for one week. A total of 142 fungal isolates were thus collected. By sequence analysis of the internal transcribed spacer region, 23 fungal genera including one unidentified taxon were found to be associated with T. castaneum. The genus Aspergillus spp. (28.9%) was the most frequently present, followed by Cladosporium spp. (12.0%), Hyphopichia burtonii (9.2%), Penicillium spp. (8.5%), Mucor spp. (6.3%), Rhizopus spp. (5.6%), Cephaliophora spp. (3.5%), Alternaria alternata (2.8%) and Monascus sp. (2.8%). Less commonly identified were genera Fusarium, Nigrospora, Beauveria, Chaetomium, Coprinellus, Irpex, Lichtheimia, Trichoderma, Byssochlamys, Cochliobolus, Cunninghamella, Mortierella, Polyporales, Rhizomucor and Talaromyces. Among the isolates, two known mycotoxin-producing fungi, Aspergillus flavus and Fusarium spp. were also identified. This result is consistent with previous studies that surveyed fungal and mycotoxin contamination in rice from RPCs. Our study indicates that the storage pest, T. castaneum, would play an important role in spreading fungal contaminants and consequently increasing mycotoxin contamination in stored rice.
The control of pests in stored grains is as economically important as increasing the crop yield because, unlike crop damage during the growing season, post-harvest damage of stored grains is not financially compensated. Fungi and animal pests are the major culprits for damage of stored grains, globally estimated to be responsible for 20% of food losses and up to 40-50% in some developing countries.
The red flour beetle (Tribolium castaneum Herbst) is one of the most important pests for stored grains such as rice (Kim and Ryoo, 1982), maize (LeCato and Flaherty, 1973), millet (Roorda et al., 1982), sorghum (Shazali and Smith, 1986), and wheat flour (Birch, 1945; Daniels, 1956) worldwide. Furthermore, T. castaneum beetles cause additional damage by spreading and promoting fungal contamination (Karunakaran et al., 2004; Kim and Ryoo, 1982; Simpanya et al., 2001). Here we chose to investigate the fungal contaminants disseminated by T. castaneum.
The T. castaneum has been reported to increase the moisture and temperature of stored grains to create an environment favorable for fungal proliferation, thereby accelerating grain degradation and decay (Miller, 1995). Degradation of stored grain by fungi results in lower germination rate, weight loss, loss of nutrients, odor and discoloration, which reduce overall grain quality. Fungal contamination of stored grains not only results in enormous economic losses but also has harmful consequences on human health and livestock due to toxic fungal secondary metabolites called mycotoxins (Tipples, 1995).
T. castaneum has also been reported to act as a vector for these toxigenic fungi during storage (Philip and Throne, 2010). When maize flour is co-contaminated with T. castaneum, toxigenic fungi including Aspergillus spp. are approximately 5 times more abundant than in the absence of the beetle vectors (Simpanya et al., 2001). Unlike in other stored grains such as wheat, barley, and corn, insectmediated fungal toxin contamination in stored rice has yet to be reported (Tanaka et al., 2004).
Aspergillus spp., which produces aflatoxin in contaminated rice, has been reported to occur mainly in high temperature and high humidity countries such as India, China, and Iran (Rahmania et al., 2011; Reddy et al., 2008). It has been reported that deoxynivalenol (DON), nivalenol (NIV), zearalenone (ZEA) and fumonisin (FMS), which are mycotoxins of genus Fusarium, were detected in stored rice (Abbas et al., 1998; Lee et al., 2011; Tanaka et al., 2004). Previously, the distribution of toxigenic fungi on rice was investigated in the southern and central regions of Korea and ochratoxin A (OTA), aflatoxin B1 (AFB1), fumonisin B1 (FB1), and zearalenone (ZEN) were detected (Park et al., 2005). According to other reports, which tested the geographic distribution of toxigenic fungi contaminating seven different types of rice samples (paddy, husk, brown, bluetinged, discolored, Broken and polished)from rice processing complexes, Fusarium spp. and Alternaria spp. were common in the southern region, while Aspergillus spp. and Penicillium spp. were common in the central region of Korea (Son et al., 2011).
Most studies previously conducted in Korea have focused on the regional distribution of contaminating fungal species, and only a few studies have examined the effect of T. castaneum on fungal transmission (Kim and Ryoo, 1982). The purpose of this study was to investigate what type of fungi could be disseminated by T. castaneum collected at rice processing complexes by time periods. This study could be used as a reference for establishing a system to effectively protect stored agricultural products.
T. castaneum beetles were collected in 49 different rice processing complexes (RPCs) nationwide, over three collection dates between April 2016 and August 2017 (Fig. 1A). We installed three or four corrugated traps (300 ├Ś 300 ├Ś 2 mm) at each RPC. Traps were placed in a variety of positions on the grain surface and collected a week later, placed individually into plastic bags. In the laboratory, the T. castaneum adults from the trap were placed into an insect breeding box (72 ├Ś 72 ├Ś 100 mm) and stored until just before the experiment.
The boxes containing T. castaneum were transferred to ŌłÆ15┬░C for 30 min (Fields, 2012), then five T. castaneum individuals were each placed on a sterilized glass slide inside a 9 cm Petri-dish lined with a single-layer of wet filter paper. The plates were incubated at 27┬░C for 7 days. After wet processing, the beetles were transferred onto potato dextrose agar (PDA) media containing streptomycin (50 mg/L) or Fusarium selective media (Nash and Snyder, 1962), and incubated at 25┬░C for 3 to 7 days. The fungal isolates were transferred to PDA medium and identified according to microscopic observations following the taxonomic keys for each genus (Barnett and Hunter, 1972; Samson et al., 1995). All fungal isolates were deposited at the Center for Fungal Genetic Resources (CFGR) at Seoul National University, Seoul, Korea.
For molecular identification, fungal genomic DNA was extracted from mycelia using DNeasy Plant Mini Kit according to the manufacturerŌĆÖs protocol (Qiagen, Valencia, CA, USA). Using the purified DNA from the collected isolates, the internal transcribed spacer with 5.8 s rDNA was amplified using ITS5/ITS4 (White et al., 1990). For further identification, beta-tubulin, calmodulin, translation elongation factor1 and glyceraldehyde-3-phosphate dehydrogenase sequence data were amplified using primer pairs BT2A/BT2B (Glass and Donaldson, 1995; OŌĆÖDonnell and Cigelnik, 1997), CL1/CL2A (OŌĆÖDonnell et al., 2000), 728F/1569R or 728F/EF2 (Carbone and Kohn, 1999; OŌĆÖDonnell and Cigelnik, 1997), and GDF1/GDR1 (Guerber et al., 2003), respectively.
PCR reactions were performed using AccuPower PCR Premix (Bioneer, Korea) with an initial denaturation for 5 min at 94┬░C, 30 cycles of 1 min denaturation at 94┬░C, 1 min annealing at 55┬░C, 1 min extension at 72┬░C, followed by a final extension for 5 min at 72┬░C. PCR products were confirmed by gel electrophoresis, purified with AccuPower PCR purification kit (Bineer, Korea) and bi-directionally sequenced on both strands with the same primers used for PCR amplification. Sequence assembly was performed using SeqMan program of DNA star (Madison, WI). The obtained nucleotide sequences were used for BLASTn search in the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/).
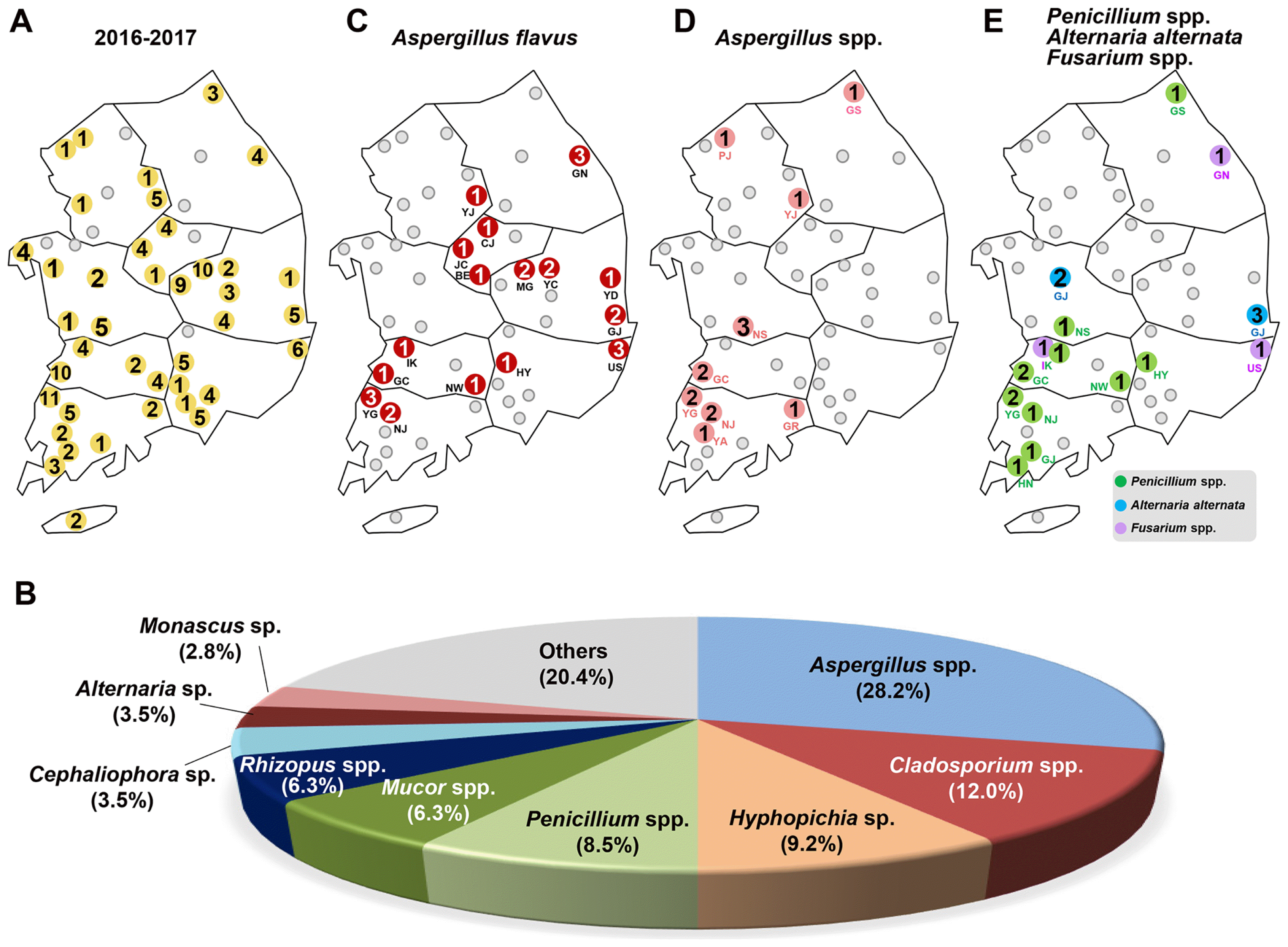
In the first round of collection (June 4, 2016), 44 fungal strains were obtained from 17 RPCs (Fig. 1B), 46 were collected in the second round from 22 RPCs (May 18, 2017) (Fig. 1C), and 52 in the third round from 26 RPCs (Aug 1, 2017) (Fig. 1D). Based on the NCBI BLAST search results of the ITS sequences and morphological analysis, Aspergillus spp. including A. flavus were dominant in whole collected periods (Fig. 1E-G).
A total 142 fungal isolates corresponding to 49 species, belonging to 23 genera, were identified from 40 RPCs (Fig. 2A, Table 1 and Table 2). The major fungal species isolated in each sampling period were Aspergillus spp. including A. flavus (40.9%), Cladosporium sp. (15.9%), and Mucor spp. (9.1%) in the first round (June 2016) (Fig. 1E), Aspergillus spp. including A. flavus (28.3%), Cladosporium sp. (17.4%), and Penicillium spp. (10.9%) in the second round (May 2017) (Fig. 1F), and Aspergillus spp. including A. flavus (19.2%), Hyphopichia sp. (15.4%), Mucor spp. (7.7%), and Penicillium spp. (7.7%) for the third round (August 2017).
The major fungal species in whole periods were Aspergillus spp. including A. flavus (28.2%), Cladosporium spp. (12.0%), Hyphopichia burtonii (9.2%), Penicillium spp. (8.5%), Mucor spp. (6.3%), Rhizopus spp. (6.3%), Cephaliophora tropica (3.5%), Alternaria alternata (3.5%), and Monascus sp. (2.8%) (Fig. 2B). Less commonly identified were Fusarium spp., Beauveria bassiana, Chaetomium globosum, Coprinellus sp., Irpex lacteus, Lichtheimia spp., Trichoderma spp., Byssochlamys spectabilis, Cochliobolus miyabeanus, Cunninghamella echinulata, Nigrospora oryzae, Mortierella oligospora, Polyporales sp., Rhizomucor pusillus, Talaromyces sp. and unidentified fungus. Among the isolates, two known mycotoxin-producing fungi, Aspergillus flavus (26 isolates) and Fusarium spp. (3 isolates) were identified.
The above results are consistent with a study which demonstrated that Aspergillus flavus is the major fungal contaminants of stored wheat in the presence of T. castaneum (Bosly and El-Banna, 2015). It is also consistent with another study on stored maize, where 10 species of fungi, Alternaria alternata, Aspergillus flavus, Aspergillus sp., Cladosporium sphaerospermum, Fusarium sp., Fusarium oxysporum, Penicillium sp., Mucor sp., Mucor racemosus and Rhizopus oryzae were isolated in the presence of T. castaneum (Simpanya et al., 2001).
In this study, we found the producer of aflatoxin, Aspergillus flavus, associated with T. castaneum, collected from16 RPCs (Gangneung (GN) in Gangwon (GW) province, Yeoju (YJ) in Gyeongii, Jincheon (JC), Chungju (CJ), and Boeun (BE) in Chungbuk (CB), Yeongdeok (YD), Gyeongju (GJ), Yecheon (YC) and Mungyeong (MG) in Gyeongbuk, Ulsan (US) and Hamyang (HY) in Gyeongnam (GN), Iksan (IK), Gochang (GC) and Namwon (NW) in Jeonbuk (JB), Yeonggwang (YG) and Naju (NJ) in Jeonnam) (Fig. 2C).
Other Aspergillus species were also found on beetles from 9 RPCs (Goseong (GS) in Gangwon province, Paju (PJ) and YJ in Gyeongii, Nonsan (NS) in Chungnam, GC in Jeonbuk, YG, NJ, Yeongam (YA) and Gurye (GR) in Jeonnam) (Fig. 2D, Table 1 and Table 2).
In addition, genus Penicillium, which is known to produce ochratoxin, was also isolated from the beetles collected in 10 RPCs (GS in Kangwon province, NS in Chungnam, Iksan (IK), GC, NS in Jeonbuk, HY in Gyeongnam, YG, NJ, Gangjin (GJ), and Haenam (HN) in Jeonnam). Another toxigenic genus, Alternaria alternata (Ostry, 2008), was found in Gongju (GJ) in Chungnam and Gyeongju (GJ) in Gyeongbuk province. Only three Fusarium species including Fusarium equiseti (Gangneung (GN) in Gangwon province), Fusarium oxysporum (Iksan (IK) in Jeonbuk), and Fusarium sp. (Ulsan (US) from Gyeongnam) were collected in 3 RPCs (Fig. 2E, Table 1). Other fungi were identified as saprophiles that proliferate on wood and debris in the facility.
The fungi Aspergillus spp., Penicillium spp., Fusarium spp., and Alternaria spp. are the major fungal species found in stored grains (Lee et al., 2011; Lee et al., 2014). More than 25% of stored grains worldwide have been reported to be contaminated with mycotoxins produced by these fungal species, and over 300 fungal metabolites have been reported to have toxicity on humans and animals (Galvano et al., 2001).
The genera Fusarium and Alternaria are known to mainly infect ears of cereal plants in the field, whereas the genera Aspergillus and Penicillium are contaminants of stored seeds, grains, and processed foods and produce mycotoxins (Adams, 1977). In particular, a number of harmful mycotoxins, such as deoxynivalenol (DON) and nivalenol (NIV), produced by Fusarium spp., and Aflatoxin produced by Aspergillus spp. are detected in stored grains (Lee et al., 2011; Lee et al., 2014; Son et al., 2011).
Both Aspergillus flavus and Fusarium spp. are known to produce mycotoxins but only Aspergillus flavus was found in this study. It is known that pests and fungi tend to co-occur in stored grains (Simpanya et al., 2001). It is necessary to investigate the distribution of pests and fungi in grain warehouses because pests promote the growth and propagation of fungi.
According to the studies on fungal and mycotoxin contamination of RPC grain samples, Aspergillus and Penicillium species were infrequently found nationwide but were particularly abundant in a few RPC samples (Lee et al., 2014). Alternaria, Nigrospora, and Epicoccum species were more consistently isolated at similar frequencies, whenever fungal contamination was detected. In accordance with the results from previous studies (Lee et al., 2014; Son et al., 2011), genera Aspergillus, Penicillium, Alternaria, and Nigrospora were identified from the T. castaneum collected at RPCs. Therefore, it is suspected that the red flour beetles are a potential vector for the transfer of toxigenic fungi and mycotoxins.
According to the study on mycotoxin contamination in different growth stages of rice (Nakaijima et al., 2008; Nash and Snyder, 1962), rice plants are always exposed to fungi and mycotoxins even before storage. So far, it has been reported that differences in temperature and humidity depending on the climate have a great influence on the growth of fungi and occurrence of mycotoxins (Russell et al., 2010). However, studies on the effect of temperature and humidity on pest-assisted mycotoxin production in stored grains are uncommon and remained to be investigated in the future. Our study shows that the storage pest, T. castaneum, could play an important role in transmission of fungi in stored rice in RPC and potentially contribute to mycotoxin contamination of rice.
Acknowledgment
This research was carried out through ŌĆ£Inventory and monitoring of biological pathogens-carrying wildlife pests for safety management of agricultural productsŌĆØ (Project Code PJ01085904) supported by Rural Development Administration, South Korea.
Fig.┬Ā1
Collation map of fungal isolates from the insect pest Tribolium castaneum in rice processing complexes (RPCs) during 2016 to 2017 in Korea. (A) Distribution map of 49 RPCs in Korea. The 49 RPCs are indicated by gray circles. The name of RPCs are noted by two capitalized letter just below the gray circles. The blue-colored letter indicates 8 provinces in Korea and two capitalized letters in parentheses indicates the abbreviation of provinces (See Table 1). The obtained fungal isolates and distribution map from (B) the first round (44 isolates from 17 RPCs), (C) the second round (46 isolates from 22 RPCs), and (D) the third round (52 isolates from 26 RPCs) of collection. The yellow colored circles indicates the location that obtained fungal isolates from the collected T. castaneum. A total number of fungal isolates is noted in the yellow colored circles. The percentage distribution of different fungal isolates from (E) the first round, (F) the second round, and (G) the third round of collection.

Fig.┬Ā2
Summary of fungal isolates from Tribolium castaneum. (A) Distribution map of 142 fungal isolates were identified from 40 RPCs. (B) The percentage distribution of different fungal isolates among the collected 142 isolates between April 2016 and August 2017. Distribution map of (C) Aspergillus flavus (collected from 12 RPCs), (D) Aspergillus spp. (16 RPCs) (E) Penicillum spp. (9 RPCs), Alternaria alternate and Fusarium spp. were collected from T. castaneum in this study. The number in the circles indicates a total number of identified isolates.
Table┬Ā1
Fungal isolates from Tribolium castaneum, GenBank accessions nos. of the ITS region sequences and the Blast search results of the sequences obtained
Table┬Ā2
Identification of four geni including Aspergillus spp., Penicillium spp., Alternaria sp. and Fusarium spp. using partial beta-tubulin, calmodulin, tef1 and glyceraldehyde-3-phosphate gene sequences
References
Abbas, HK, Shier, WT, Seo, JA, Lee, YW and Musser, SM 1998. Phytotoxicity and cytotoxicity of the fumonisin C and P series of mycotoxins from Fusarium spp. fungi. Toxicon. 36:2033-2037.


Adams, JM 1977. A review of the literature concerning losses in stored cereals and pulses. Trop Sci. 19:1-7.
Barnett, HL and Hunter, BB 1972. Illustrated Genera of Imperfect Fungi. 3rd ed. Burgess Publishing Co, Minneapolis, Minnesota, USA. 241.
Birch, LC 1945. The influence of temperature, humidity and density on the oviposition of the small strain of Calandra oryzae L. and Rhizopertha dominica Fab (Coleoptera). Aust J Exp Biol Med Sci. 23:197-203.


Bosly, HA and El-Banna, OM 2015. Isolation and identification of fungal growth on Tribolium castaneum in stored wheat flour. J Entomol Nematol. 7:11-17.

Carbone, I and Kohn, LM 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 91:553-556.

Daniels, NE 1956. Damage and reproduction by the flour beetles, Tribolium confusum and T. castaneum in wheat at three moistures. J Econ Entomol. 49:244-247.

Fields, P, Bhadriraju, S and Hulasare, R 2012. Extreme temperatures. In: Stored Product Protection, eds. by DW Hagstrum, TW Philip and G Cuperus, 179-190. K-State Research and Extension, Manhattan, USA.
Galvano, F, Piva, A, Ritieni, A and Galvano, G 2001. Dietary strategies to counteract the effects of mycotoxins: a review. J Food Prot. 64:120-131.


Glass, L and Donaldson, GC 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 61:1323-1330.



Guerber, JC, Liu, B, Correll, JC and Johnston, PR 2003. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia. 95:872-895.


Karunakaran, C, Jayas, DS and White, NDG 2004. Identification of wheat kernels damaged by the red flour beetle using X-ray images. Biosyst Eng. 87:267-274.

Kim, YB and Yu, MI 1982 Activities of molds and insects during rice storage Part I. Activities of Tribolium castaneum Herbst and Aspergillus species. J Korean Agric Chem Soc. 25:252-256 (in Korean).
LeCato, GL and Flaherty, BR 1973. Tribolium castaneum progeny production and development on diet supplemented with egg of adult of Plodia interpunctella. J Stored Prod Res. 9:199-203.

Lee, T, Lee, SH, Lee, SH, Shin, JY, Yun, JC, Lee, YW and Ryu, JG 2011. Occurrence of Fusarium mycotoxins in rice and its milling by-products in Korea. J Food Prot. 74:1169-1174.


Lee, T, Lee, S, Kim, LH and Ryu, JG 2014 Occurrence of fungi and Fusarium mycotoxins in the rice samples from rice processing complex. Res Plant Dis. 20:289-294 (in Korean).

Miller, JD 1995. Fungi and mycotoxins in grain: implications for stored product research. J Stored Prod Res. 31:1-16.

Nakaijima, T, Yoshida, M and Tomimura, K 2008. Effect of lodging on the level of mycotoxins in wheat, barley, and rice infected with the Fusarium graminearum species complex. J Gen Plant Pathol. 74:289-295.

Nash, SM and Snyder, WC 1962. Quantitative estimations by plate counts of propagules of the bean root rot Fusarium in field soils. Phytopathology. 52:567-572.
OŌĆÖdonnell, K and Cigelnik, E 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 7:103-116.


OŌĆÖdonnell, K, Nirenberg, HI, Aoki, T and Cigelnik, E 2000. A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience. 41:61-78.

Ostry, V 2008. Alternaria mycotoxins: an overview of chemical characterization, producers, toxicity, analysis and occurrence in foodstuffs. World Mycotoxin J. 1:175-188.

Park, JW, Choi, SY, Hwang, HJ and Kim, YB 2005. Fungal mycoflora and mycotoxins in Korean polished rice destined for humans. Int J Food Microbiol. 103:305-314.


Philip, TW and Throne, JE 2010. Biorational approaches for managing stored-product insect. Annu Rev Entomol. 55:375-397.


Rahmani, A, Soleimany, F, Hosseini, H and Nateghi, L 2011. Survey on the occurrence of aflatoxins in rice from different provinces of Iran. Food Addit Contam B. 4:185-190.

Reddy, KRN, Reddy, CS, Abbas, HK, Abel, CA and Muralidharan, K 2008. Mycotoxigenic fungi, mycotoxins, and management of rice grains. Toxin Rev. 27:287-317.

Roorda, FA, Schulten, GGM and Andriessen, EAM 1982. Laboratory observations on the development of Tribolium castaneum Herbst (Col., Tenebrionidae) on millet at different temperatures and relative humidities. J Appl Entomol. 93:446-452.

Paterson, RRM and Lima, N 2010. How will climate change affect mycotoxins in food? Food Res Int. 43:1902-1914.

Samson, RA and De Boer, E 1995. Introduction to food-bornd fungi. 4th ed. Centraalbureau voor Schimmelcultures, Baarn, Netherlands. 299.
Shazali, MEH and Smith, R 1986. Life history studies of externally feeding pest of stored sorghum: Corcyra cephalonica (Staint.) and Tribolium castaneum (Hbst). J Stored Prod Res. 22:55-61.

Simpanya, MF, Allotey, J and Mpuchane, S 2001. Insect and mycoflora interactions in maize flour. Afr J Food Nut Sci. 1:3-8.
Son, SW, Nam, YJ, Lee, SH, Lee, SM, Lee, SH, Kim, MJ, Lee, T, Yun, JC and Ryu, JG 2011 Toxigenic fungal contaminants in the 2009-harvested rice and its milling-by products samples collected from rice processing complexes in Korea. Res Plant Dis. 17:280-287 (in Korean).

Tanaka, K, Kobayashi, H, Nagata, T and Manabe, M 2004. Natural occurrence of trichothecenes on lodged and water damaged domestic rice in Japan. J Food Hyg Soc Jpn. 45:63-66.

Tipples, KH 1995. Quality and nutritional changes in stored grain. In: Stored-grain ecosystems, eds. by DS Jayas, NDG White and WE Muir, 325-351. New York Marcel Dekker, New York, USA.
- TOOLS
-
METRICS

- Related articles
-
Isolation and Identification of Antifungal Fatty Acids from the Extract of Common1986 August;2(2)



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print



