Rapid and Specific Detection of Apple stem grooving virus by Reverse Transcription-recombinase Polymerase Amplification
Article information
Abstract
Apple stem grooving virus (ASGV) is considered to cause the most economically important viral disease in pears in Korea. The current PCR-based methods used to diagnose ASGV are time-consuming in terms of target detection. In this study, a novel assay for specific ASGV detection that is based on reverse transcription-recombinase polymerase amplification is described. This assay has been shown to be reproducible and able to detect as little as 4.7 ng/μl of purified RNA obtained from an ASGV-infected plant. The major advantage of this assay is that the reaction for the target virus is completed in 1 min, and amplification only requires an incubation temperature of 42°C. This assay is a promising alternative method for pear breeding programs or virus-free certification laboratories.
Body
The Asian pear [Pyrus pyrifolia (N. L. Burm.) Nakai] is one of the main fruit trees in Korea. Apple stem grooving virus (ASGV) occurs frequently in single or mixed infections and causes a significant yield reduction in pear cultivars (Cho et al., 2010). ASGV, which causes black necrotic spot disease, decreases the growth and productivity of pear trees (Lee et al., 2017). In addition, ASGV, a member of the genus Capillovirus is associated with latent infections in many commercial apple and pear cultivars (Adams et al., 2012). ASGV infections are symptomless, however, some ASGV strains can induce necrotic spot vein yellow diseases in pear trees.
Virus detection is a crucial step to guarantee the virus-free certificate in fruit trees. So far, serological and nucleic acid-based assays are the most widely used methods for detection methods of plant viruses (Jeong et al., 2014; Ji et al., 2013; Yao et al., 2014). However, these methods are time-consuming (3–4 h) and require expensive equipment and trained personnel. Recently, isothermal amplification methods (for example, loop-mediated isothermal amplification (LAMP)), have been developed for the detection of pear viruses, however, while this provides a simple and rapid detection system for diagnoses for pear viruses in the field and in poorly equipped laboratories, it requires four to six DNA oligos as well as more difficult downstream analysis such as cloning and direct sequencing (Lu et al., 2018; Zhao et al., 2014).
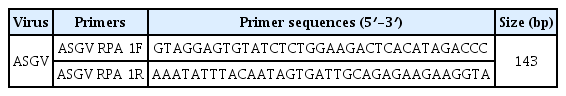
Recombinase polymerase amplification (RPA) is another type of isothermal amplification method that targets and amplifies DNA from samples (Piepenburg et al., 2006). The RPA method uses DNA-binding proteins (two ATP-dependent recombinases, [usvX and usvY] and single-strand DNA binding protein [called gp32]) which bind the DNA oligos, and scan for the homologous sequence (target) and a mesophilic polymerase (Bacillus subtilis DNA polymerase І), which extends the 3′ end of the invading DNA oligos using the opposite strand as a template. The RPA product is amplified at a constant low temperature (ranging from 25°C to 42°C) in less than 30 min. Gel electrophoresis and probe-based fluorescence are one of the most commonly used methods to detect RPA products. In the case of RNA targets, for example plant RNA viruses, a reverse transcriptase can be included in the RNA reactions (Daher et al., 2016). Although RT-RPA technology has been used for the detection of other plant viruses such as Little cherry virus 2, Plum pox virus, and Yam mosaic virus because of its superior speed, portability, and accessibility, it has rarely been applied to pear viruses (Londoño et al., 2016; Mekuria et al., 2014; Silva et al., 2015; Zhang et al., 2014). In this study, a novel RT-RPA method was developed for the detection of ASGV, a pear virus. This assay offers advantages that can enable the rapid screening of pear virus-free materials for further propagation and distribution to farmers. To evaluate the distribution of ASGV in Korea, 158 leaf samples exhibiting typical symptoms, including leaf chlorosis and necrotic spots were collected from 34 pear orchards in five provinces in Korea (Sangju, Namyangju, Ulsan, Chonan, and Naju). Total RNA was extracted from the leaf tissue using the IQeasy plus plant RNA extraction Mini Kit (iNtRON, Korea) according to the manufacturer’s recommendations. The majority of positive samples from trees showing ASGV symptoms and negative samples (ASGV-free) were confirmed by RT-PCR for the presence of ASGV using diagnostic primers (Cho et al., 2010) and stored at −80°C for use as RT-RPA assay templates. One-step RT-PCR was performed using the materials and protocols in the SuPrimeScript RT-PCR Premix (GeNet Bio, Daejeon, Korea). The RT-RPA products of the complete coat protein (CP) coding sequences amplified were determined after cloning into pCR2.1-TOPO TA vector (Invitrogen, Carlsbad, Grand Island, NY) and clones were sequenced. Four ASGV isolates (GenBank accession No. MG682507, MG682506, MG682508, and MG682509) were deposited to NCBI. To design the primers for the RT-RPA assays, the sequences of 15 ASGV isolates including Korea isolates were examined to identify the conserved regions in the virus genome. The CP gene sequences of ASGV (GenBank accession No. MG682507, MG682506, MG682508, MG682509, JN792471, LC084659, KX988001, KF735124, LC143387, KR185346, KU198289, LC184611, AB004063, KJ579253, and GQ330294) were then aligned (Fig. 1A). The highly-conserved regions of ASGV-CP were selected and primers were designed according to the TwistDx RPA instruction manual (TwistDx, Ltd., Cambridge, UK) (Table 1). The primers were designed based on the nucleotide position 43–185 from the genomic RNA CP gene, generating a 143 bp product. Several combinations of DNA oligos were initially evaluated for target sequences and only one product was generated by RT-RPA because of the non-specific amplification associated with RT-RPA (data not shown).
Primer position within the coat protein of Apple stem grooving virus (ASGV) and the detection of ASGV by RT-RPA. (A) Clustal W multiple sequence alignment was performed with Bio-edit using the sequence of Korea ASGV isolates with isolates from other countries. Unfilled boxes represent the primer regions used in this study. (B) RT-RPA amplification products of ASGV. M, DNA marker; lane 1, ASGV-infected tissues; lane 2, non-infected tissues control. Five independent reactions were performed, and similar results were obtained.
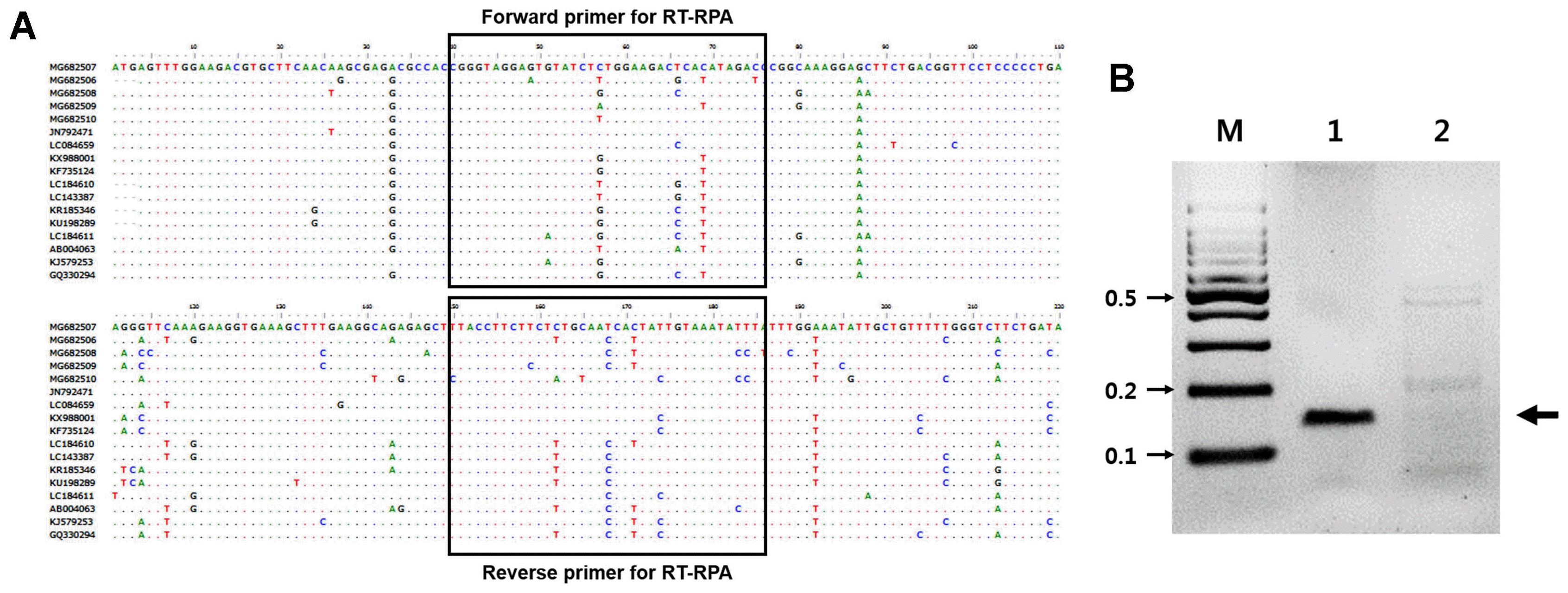
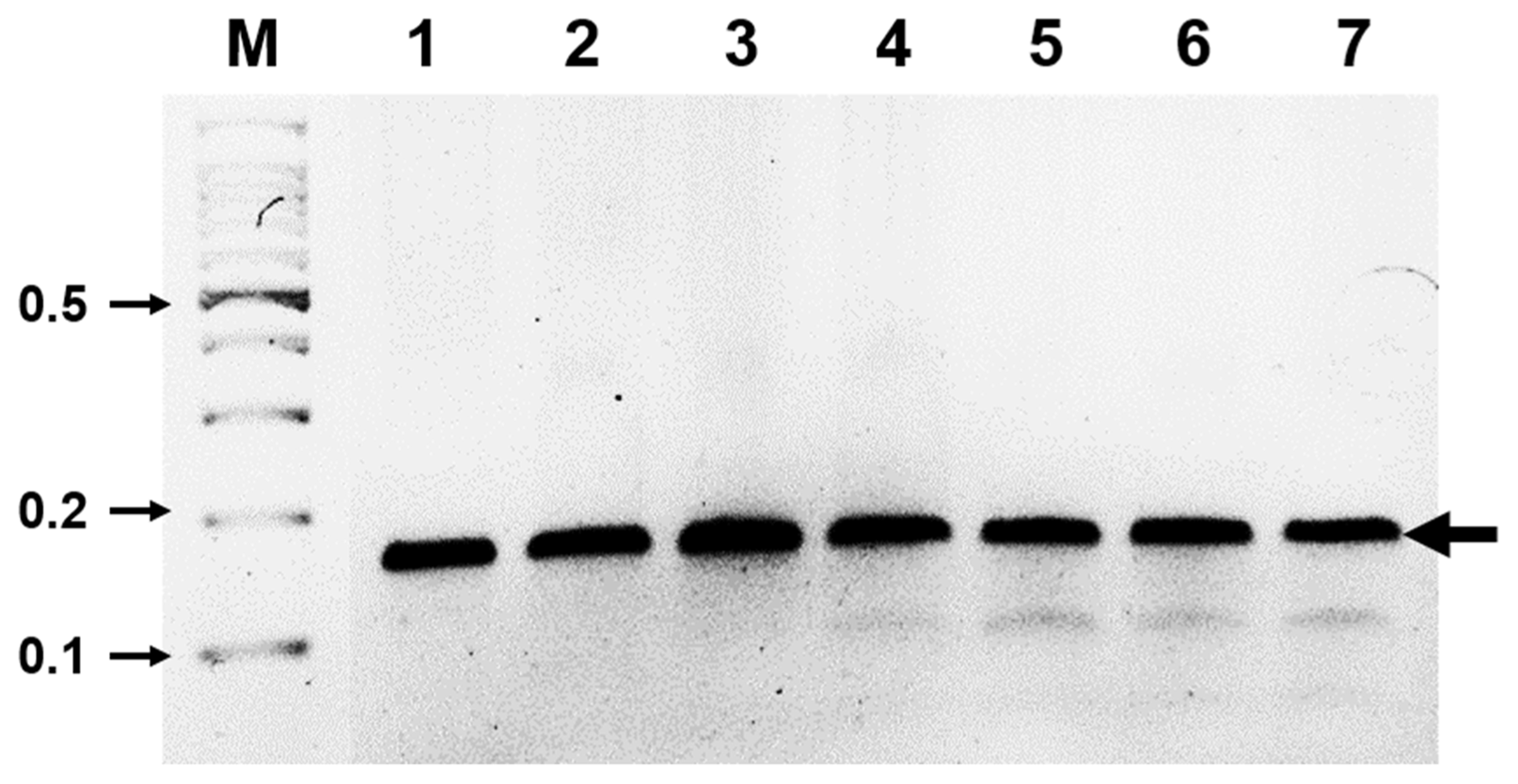
RT-RPA assays were performed using the Twist Amp Basic RT kit reagents (TwistDx Limited) in a 50 μl total volume according to the manufacturer’s instructions. This kit provides a dried enzyme pellet to reaction microtubes. The mixture contained 10 μM of each RT-RPA primer, 280 mM of magnesium acetate, and 1 μl (47 ng/μl; total RNAs from ASGV-infected tissue) of template. The reactions were then incubated at 42°C for 30 min using a water bath. Next, the RT-RPA products were analyzed in a 3% agarose gel containing ethidium bromide (EtBr). As shown in Figure 1B, RT-RPA products were clearly detected with the expected size (143 bp). Only a very low amplification background was present in the non-infected tissues of the pear leaves. No amplicons of the expected size were generated from the RNA extracted from non-infected leaves (Fig. 1B). The RT-RPA assays could also detect ASGV from pear fruits and pear tree seedlings (Supplementary Fig. 1). The identity of the amplicons was confirmed through cloning and sequencing. Of the 30 samples that tested positive by RT-PCR, all 30 samples were positive when assayed by RT-RPA. Of the 10 samples that tested negative by RT-PCR, all 30 were also negative when tested by RT-RPA (data not shown). The overall diagnostic coincidence between RT-RPA and RT-PCR was 100%. To optimize the reaction time, the ASGV-infected total RNAs were amplified for 1, 3, 5, 10, 15, 20, or 30 min. The amplified products were electrophoresed on a 3% (W/V) agarose gel and subsequently stained with EtBr. As shown in Fig. 2, RT-RPA products were clearly visible, even with a short reaction time (1 min). However, there was no significant difference in product yield from 1 min to 30 min reactions, suggesting that after the 1 min reaction, the dNTPs or other enzymatic components in the reaction were completely used up. The detection limit of the RT-RPA assay was ascertained by testing 10-fold serial dilutions (starting from 47 ng/μl) of the purified RNA obtained from the ASGV-infected pear leaves, and through comparison with RT-PCR results; 1 μl of each RNA dilution was amplified by RT-RPA as well as by conventional RT-PCR. The RT-PCR and RT-RPA assays used the same primers. 1 μl of template was mixed with each virus-specific primer set and the SuprimeScript RT-PCR Premixture (GeNet Bio, Daejeon, Korea). RT-PCR was conducted as follows: stage 1, 50°C for 30 min; stage 2, 95°C for 5 min; stage 3, 35 cycles of 95°C for 30 s, 56°C for 30 s and 72°C for 40 s; and stage 4, 72°C for 5 min. The PCR products were analyzed through agarose gel electrophoresis. A representative assay is shown in Fig. 3. Consistently positive results were obtained in the RT-RPA for ASGV-infected samples containing at least 4.7 ng/μl of RNA (10−1 dilution) (Fig. 3). In the RT-PCR assays, positive results were obtained for the ASGV-infected samples containing at least 470 pg/μl of RNA (10−2 dilution) (Fig. 3B). Five independent reactions were performed with similar results observed in all five. These results indicate that the level of sensitivity of the RT-RPA method for ASGV detection was less sensitive than that of RT-PCR. Although these methods exhibited comparable sensitivities, the expertise and time required to conduct an RT-RPA assay is significantly reduced (1–30 min) compared to those required to perform RT-PCR (2 h). These results demonstrate that both the RT-RPA and RT-PCR methods possess comparable sensitivity for the detection of ASGV. It is known that low-temperature amplification usually creates a high background signal; hence, the non-specific bands that appeared during the RT-RPA are not uncommon (Londoño et al., 2016). However, non-specifically amplified bands can be easily differentiated based on their sizes and signal intensities (Fig. 3). The specificity of the RT-RPA assay was confirmed by cross reactions assays using RNAs from Apple chlorotic leafspot virus (ACLSV) and Apple stem pitting virus (ASPV), distinct viruses that are often present in pears in Korea, particularly through-infected pear leaves. RT-RPA did not amplify the templates of other important pear viruses, indicating a high specificity of the primers used (Fig. 4). DNA oligos used for RT-RPA in this study were derived from conserved sequences of field-collected ASGV Korean isolates, so the developed RT-RPA assay can be applied to the detection of a wide spectrum of ASGV isolates in Korea.
Determination of optimal reaction time for ASGV diagnosis using RT-RPA. ASGV was amplified with RT-RPA for different time points and a clear DNA band of the expected size (143 bp) could be visualized by agarose gel electrophoresis. M, DNA marker; lanes 1–7, DNA products from reactions incubated for 1, 3, 5, 10, 15, 20, and 30 min, respectively.
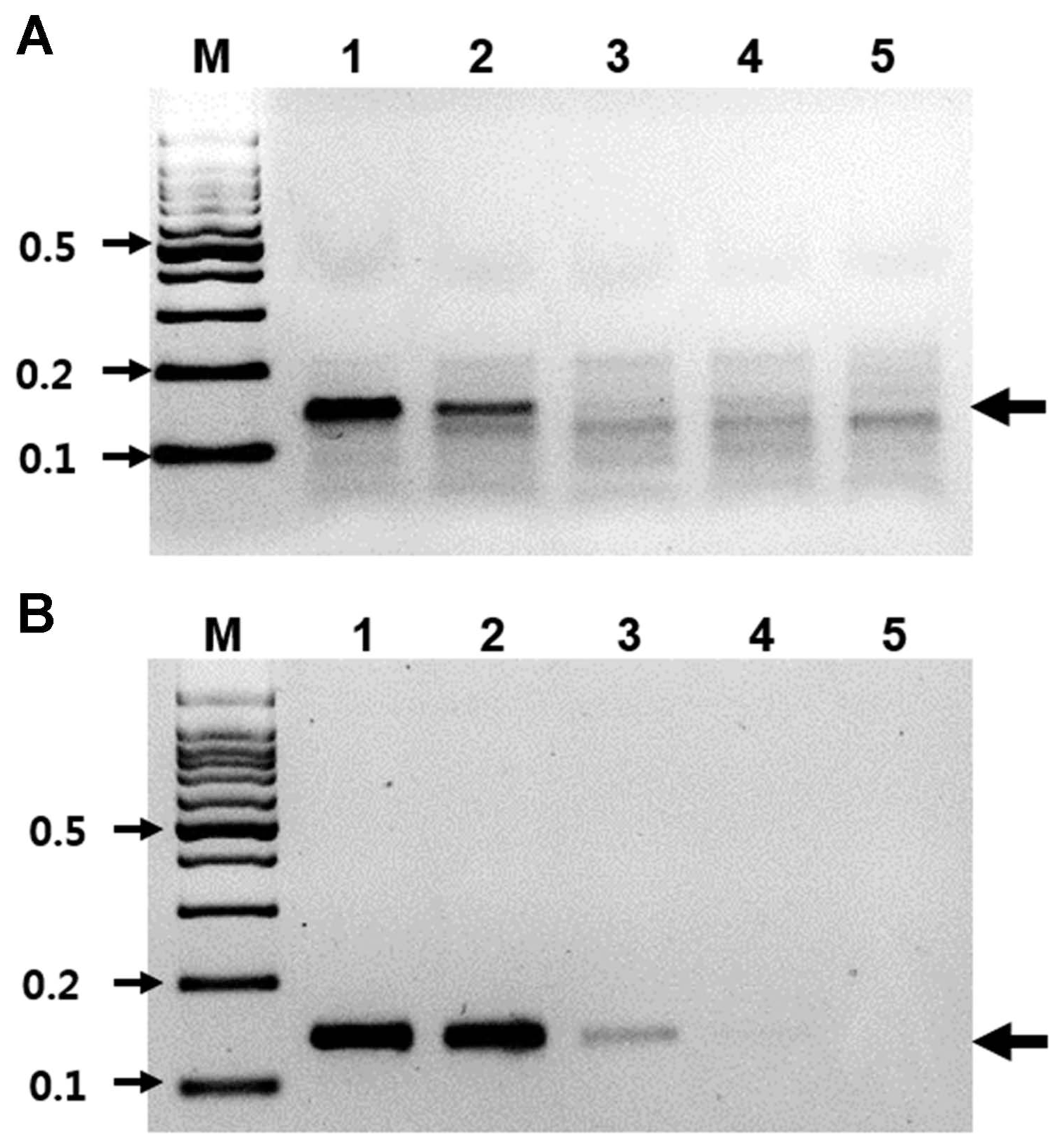
Sensitivity of RT-RPA. (A) The detection limit of the RT-RPA assay using total RNA isolated from pear leaves infected with ASGV. M, DNA marker; lanes 1–5, serial 10-fold dilutions of RNA (ranging from 47 ng/μl to 4.7 pg/μl). (B) The detection limit of the RT-PCR assay using total RNA isolated from pear leaves infected with ASGV. M, DNA marker; lanes 1–5, serial 10-fold dilution of RNA (ranging from 47 ng/μl to 4.7 pg/μl).

Specificity of RT-RPA for ASGV. Total RNAs from the ASGV-, ACLSV-, and ASPV-infected tissues were all tested using RT-RPA. M, DNA marker; lane 1, ASGV-infected tissues; lane 2, ACLSV-infected tissues; lane 3, ASPV-infected tissues; lane 4, virus-free tissues; lane 5, no-template virus. Five independent reactions were performed with similar results observed for all five reactions.
ASGV were detected using one-step real-time RT-PCR from apple trees (Winkowska et al., 2016). However, one-step RT-qPCR assays are long (approximately 100 min) and require expensive and specific equipment. Rapid and specific detection of ASGV using reverse transcription (RT)-LAMP has also been reported for apple and pear leaves (Lu et al., 2018). However, RT-LAMP assays require four or six primers and more difficult downstream analysis, such as cloning and direct sequencing, limitations that are overcome by RPA. The short reaction time combined with the requirement of a temperature in the same region of 42°C indicates that the RT-RPA assay developed in this study has several advantages over the RT-PCR currently used for ASGV diagnosis. One such advantage is that RT-RPA does not have the same temperature requirements to amplify the products. This assay, in principle, only requires a simple heating device to detect viruses. This study is the first report on the use of a novel assay based on an isothermal amplification technique, RT-RPA for the detection of pear virus. The developed assay is highly specific and rapid, finishing in 1 min. Even though the cost per sample for RT-RPA analysis is approximately $6, which is equivalent to the cost per sample for end-point RT-PCR, the reaction time required for the RT-RPA assay is shorter (Londoño et al., 2016).
In conclusion, the RT-RPA method developed in this study demonstrates higher speed and specificity for the detection of ASGV from pear leaves compared to an RT-PCR-based assay. In addition, a time-point experiment showed that RT-RPA can detect pear virus in only one minute. Moreover, the fragment of ASGV amplified by RT-RPA can be determined within two minutes in the field. The developed RT-RPA assay is expected to be used directly in commercial nurseries and diagnostic laboratories for the rapid detection of ASGV, thereby helping prevent the spread of pear viruses.
Supplementary Information
Acknowledgments
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through (Agri-Bioindustry Technology Development Program), funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (No. 317006-04-2-HD030).