Abdel-Razzak, HS, Alfrmawy, AM, Ibrahim, HM and El-Hanafy, AA 2012. Genetic diversity in faba bean (Vicia faba L.) using inter-simple sequence repeat (ISSR) markers and protein analysis. Life Sci J. 9:497-503.
Abdel Rehim, MA 1962. Studies on the organisms causing root rot and wilt of horse beans (Vicia faba var. equina) in U. A. R. PhD thesis. Faculty of Agriculture, Alexandria University, Qism Bab Sharqi, Egypt.
Abdul Wahid, OA, Ibrahim, ME and Omar, MA 1998. Occurrence of soil suppressiveness to Fusarium wilt disease of broad bean in Ismailia Governorate.
J Phytopathol. 146:431-435.

Abou-Zeid, NM, El-Morsy, GA, Hassanein, AM and Arafa, MK 1997. Major organisms causing root-rot, wilt and their relative importance on faba bean, lentil and chickpea. Egypt J Agric Res. 75:529-542.
Ahmed, HFS, El-Araby, MM and Omar, SA 2002. Differential effect of jasmonic acid on the defense of faba bean against Fusarium wilt: modulation of other phytohormones and simple phenols. Int J Agric Biol. 4:447-453.
Alghamdi, SS 2009a. Chemical composition of faba bean (Vicia faba L.) genotypes under various water regimes.
Pak J Nutr. 8:477-482.

Alghamdi, SS 2009b. Varietal identification and genetic purity assessment of F
1 hybrid seeds using RAPD markers in faba bean (Vicia faba L.).
Acta Hortic. 829:269-274.

Alghamdi, SS, Al-Faifi, SA, Migdadi, HM, Ammar, MH and Siddique, KHM 2011. Inter-simple sequence repeat (ISSR)-based diversity assessment among faba bean genotypes.
Crop Pasture Sci. 62:755-760.

Alghamdi, SS, Al-Faifi, SA, Migdadi, HM, Khan, MA, El-Harty, EH and Ammar, MH 2012. Molecular diversity assessment using sequence related amplified polymorphism (SRAP) markers in Vicia faba L.
Int J Mol Sci. 13:16457-16471.



Ali, MB, Haridy, AG and Mahmoud, AF 2019. Evaluation of faba bean genotypes for yield and resistance to Fusarium root rot under greenhouse and field conditions. Int J Biosci. 14:374-385.
Almagro, L, G├│mez Ros, LV, Belchi-Navarro, S, Bru, R, Ros Barcel├│, A and Pedre├▒o, MA 2009. Class III peroxidases in plant defence reactions.
J Exp Bot. 60:377-390.


Amar, MH and El Wahab, MA 2013. Comparative genetic study among Origanum L. plants grown in Egypt. J Biodivers Environ Sci. 3:208-222.
Ammar, MH, Alghamdi, SS, Migdadi, HM, Khan, MA, El-Harty, EH and Al-Faifi, SA 2015. Assessment of genetic diversity among faba bean genotypes using agro-morphological and molecular markers.
Saudi J Biol Sci. 22:340-350.



Balkaya, A and Yanmaz, R 2002. Morphological properties of cultivar nominate selected Black Sea Region bean populations and identification by protein markers. Ankara Univ J Agric Sci. 9:182-188.
Bebeli, P and Kaltsikes, P 1993. New developments in varietal identification. In: Seed science and technology, eds. by AJG van Gastel, MA Pagnotta and E Porceddu, 161-172. ICARDA, Aleppo, Syria.
Booth, C 1971. The genus Fusarium. Commonwealth Agricultural Bureaux (for the) Commonwealth Mycological Institute, Kew, UK. 237.
Budak, H, Shearman, RC, Parmaksiz, I, Gaussoin, RE, Riordan, TP and Dweikat, I 2004. Molecular characterization of buffalograss germplasm using sequence-related amplified polymorphism markers.
Theor Appl Genet. 108:328-334.


Colditz, F, Nyamsuren, O, Niehaus, K, Eubel, H, Braun, HP and Krajinski, F 2004. Proteomic approach: identification of Medicago truncatula proteins induced in roots after infection with the pathogenic oomycete Aphanomyces euteiches.
Plant Mol Biol. 55:109-120.


Costa, R, Pereira, G, Garrido, I, Tavares-de-Sousa, MM and Espinosa, F 2016. Comparison of RAPD, ISSR, and AFLP molecular markers to reveal and classify Orchard grass (Dactylis glomerata L.) germplasm variations.
PLoS ONE. 11:e0152972



Dhole, VJ and Reddy, KS 2013. Development of a SCAR marker linked with a MYMV resistance gene in mungbean (Vigna radiata L. Wilczek).
Plant Breed. 132:127-132.

Dice, LR 1945. Measures of the amount of ecologic association between species.
Ecology. 26:297-302.

Dong, Y, Dong, K, Zheng, Y, Tang, L and Yang, ZX 2014 Faba bean Fusarium wilt (Fusarium oxysporum) control and its mechanism in different wheat varieties and faba bean intercropping system. J Appl Ecol. 25:1979-1987 (in Chinese)..
Duc, G 1997. Faba bean (Vicia faba L).
Field Crops Res. 53:99-109.

El-Khallal, SM 2007. Induction and modulation of resistance in tomato plants against Fusarium wilt disease by bioagent fungi (arbuscular mycorrhiza) and/or hormonal elicitors (Jasmonic acid & Salicylic acid): 2-changes in the antioxidant enzymes, phenolic compounds and pathogen related- proteins. Aust J Basic Appl Sci. 1:717-732.
El-Sayed, ZG 2006. Genetic markers for some important traits in faba bean (Vicia faba L.). PhD thesis. Department of Genetics, Faculty of Agriculture, Ain Shams University, Cairo, Egypt.
Elwakil, MA, El-Refai, IM, Awadallah, OA, El-Metwally, MA and Mohammed, MS 2009. Seed-borne pathogens of faba bean in Egypt: detection and pathogenicity. Plant Pathol J. 8:90-97.
Eujayl, I, Erskine, W, Bayaa, B, Baum, M and Pehu, E 1998. Fusarium vascular wilt of lentil: inheritance and identification of DNA markers for resistance.
Plant Breed. 117:497-499.

Gerlach, W and Nirenberg, HI 1982. The genus Fusarium: a pictorial atlas.
Mitt Biol Bundesanst Land-Forstwirtsch Berlin-Dahlem. 209:1-406.

Ghafoor, A, Ahmad, Z, Qureshi, AS and Bashir, M 2002. Genetic relationship in Vigna mungo (L.) Hepper and V. radiata (L.) R. Wilczek based on morphological traits and SDS-PAGE. Euphytica. 123:367-378.
Ghislain, MD, Zhang, D, Fajardo, D, Huam├Īn, Z and Hijmans, RJ 1999. Marker-assisted sampling of the cultivated Andean potato Solanum phureja collection using RAPD markers. Genet Resour Crop Evol. 46:547-555.
Giovannelli, JL, Farnham, MW and Wang, M 2002. Development of sequence characterized amplified region markers linked to downy mildew resistance in broccoli.
J Am Soc Hortic Sci. 127:597-601.

Gomez, KA and Gomez, AA 1984. Statistical procedures for agricultural research. 2nd ed. John Wiley and Sons, New York, NY, USA. 704.
Guikema, JA and Sherman, LA 1981. Electrophoretic profiles of cyanobacterial membrane polypeptides showing heme-dependent peroxidase activity.
Biochim Biophys Acta. 637:189-201.

Gupta, VK, Misra, AK, Gaur, RK, Jain, PK, Gaur, D and Sharma, S 2010. Current status of Fusarium wilt disease of guava (Psidium guajava L.) in India.
Biotechnology. 9:176-195.

Hao, Q, Liu, Z-A, Shu, QY, Zhang, R, De Rick, J and Wang, L-S 2008. Studies on Paeonia cultivars and hybrids identification based on SRAP analysis.
Hereditas. 145:38-47.


Jhang, T, Kaur, M, Kalia, P and Sharma, TR 2010. Efficiency of different marker systems for molecular characterization of subtropical carrot germplasm.
J Agric Sci. 148:171-181.

Johnson, R and Jellis, GJ 1992. Development in plant pathology. Vol. 1. Breeding for disease resistance. Kluwer Academic, Dordrecht, Netherlands. 206.
Khalaf, AEA, Afiah, SA, Khalil, RMA and Ahmed, AIS 2015. Molecular markers for resistance of chocolate spot disease in faba bean (Vicia faba L.) using ISSR-PCR. Middle-East J Sci Res. 23:558-567.
Kamboj, RK, Pandey, MP and Chaube, HS 1990. Inheritance of resistance to Fusarium wilt in Indian lentil germplasm (Lens culinaris Medik.).
Euphytica. 50:113-117.

Kleczewski, NM and Egel, DS 2011. A diagnostic guide for Fusarium wilt of watermelon.
Plant Health Prog. 12:27

Kumari, YSMAI and Vengadaramana, A 2017. Stimulation of defense enzymes in tomato (Solanum lycopersicum L.) and chilli (Capsicum annuum L.) in response to exogenous application of different chemical elicitors.
Univers J Plant Sci. 5:10-15.

Laemmli, UK 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4.
Nature. 227:680-685.


Li, G and Quiros, CF 2001. Sequence-related amplified polymorphism (SRAP), a new marker system based on a sample PCR reaction: its application to mapping and gene tagging in Brassica.
Theor Appl Genet. 103:455-461.

Lioi, L, Piergiovanni, AR, Pignone, D, Puglisi, S, Santantonio, M and Sonnante, G 2005. Genetic diversity of some surviving on-farm Italian common bean (Phaseolus vulgaris L.) landraces.
Plant Breed. 124:576-581.

Mace, ES, Phong, DT, Upadhyaya, HD, Chandra, S and Crouch, JH 2006. SSR analysis of cultivated groundnut (Arachis hypogaea L.) germplasm resistant to rust and late leaf spot diseases.
Euphytica. 152:317-330.

Mahmoud, AF 2016a. Evaluation of certain antagonistic fungal species for biological control of faba bean wilt disease incited by Fusarium oxysporum. J Phytopathol Pest Manag. 3:1-14.
Mahmoud, AF 2016b. Genetic variation and biological control of Fusarium graminearum isolated from wheat in Assiut-Egypt.
Plant Pathol J. 32:145-156.


Mahmoud, AF, Abou-Elwafa, SF and Shehzad, T 2018. Identification of charcoal rot resistance QTLs in sorghum using association and in silico analyses.
J Appl Genet. 59:243-251.


Mahmoud, AF, Hassan, MI and Amein, KA 2015. Resistance potential of bread wheat genotypes against yellow rust disease under Egyptian climate.
Plant Pathol J. 31:402-413.



Mantel, N 1967. The detection of disease clustering and a generalized regression approach.
Cancer Res. 27:209-220.

Mazen, MM, El-Batanony, NH, Abd El-Monium, MM and Massoud, ON 2008. Cultural filtrate of Rhizobium spp. and arbuscular mycorrhiza are potential biological control agents against root rot fungal diseases of faba bean. Global J Biotechnol Biochem. 3:32-41.
Mofunanya, A and Edu, EA 2015. Physiological and biochemical changes in Cucurbita moschata Duch Ex Poir inoculated with a Nigerian strain of moroccan watermelon mosaic virus (MWMV): Lagenaria breviflora isolate.
Int J Plant Pathol. 6:36-47.

Mondal, S and Badigannavar, AM 2010. Molecular diversity and association of SSR markers to rust and late leaf spot resistance in cultivated groundnut (Arachis hypogaea L.).
Plant Breed. 129:68-71.

Murray, MG and Thompson, WF 1980. Rapid isolation of high molecular weight plant DNA.
Nucleic Acids Res. 8:4321-4325.



Nagaraja, NR, Kumar, MBA, Prasad, CTM, Rajkumar, , Sinha, P and Jain, J 2016. Genetic diversity studies and screening for Fusarium wilt (Fusarium udum Butler) resistance in wild pigeonpea accessions, Cajanus scarabaeoides (L.) Thouars.
Indian J Plant Genet Resour. 29:121-129.

Neog, B, Yadav, RNS and Singh, ID 2004. Peroxidase, polyphenol oxidase and acid phosphatase activities in the stigma-style tissue of Camellia sinensis (L) O. Kuntze following compatible and incompatible pollination. J Indian Inst Sci. 84:47-52.
Nyamsuren, O, Colditz, F, Rosendahl, S, Tamasloukht, MB, Bekel, T, Meyer, F, Kuester, H, Franken, P and Krajinski, F 2003. Transcriptional profiling of Medicago truncatula roots after infection with Aphanomyces euteiches (oomycota) identifies novel genes upregulated during this pathogenic interaction.
Physiol Mol Plant Pathol. 63:17-26.

Okiror, MA 2002. Genetics of resistance to Fusarium udum in pigeonpea (Cajanus cajan (L.) Millsp.). Indian J Genet Plant Breed. 62:218-220.
Parmar, PP, Bhatt, KN, Oza, VP, Patel, AD, Kathiria, KB and Subramanian, RB 2009. Microsatellite marker associated with fusarium wilt resistance in tomato. World J Agric Sci. 5:389-393.
Powell, W, Machray, GC and Provan, J 1996. Polymorphism revealed by simple sequence repeats.
Trends Plant Sci. 1:215-222.

Prasad, P, Eswara Reddy, NP, Anandam, RJ and Lakshmikantha Reddy, G 2003. Isozymes variability among Fusarium udum resistant cultivars of pigeonpea (Cajanus cajan (L.) (Millsp).
Acta Physiol Plant. 25:221-228.

Prevost, A and Wilkinson, MJ 1999. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars.
Theor Appl Genet. 98:107-112.

Provan, J, Thomas, WTB, Forster, BP and Powell, W 1999. Copia-SSR: a simple marker technique which can be used on total genomic DNA.
Genome. 42:363-366.

Rabey, JM, Vered, Y, Shabtai, H, Graff, E and Korczyn, AD 1992. Improvement of parkinsonian features correlate with high plasma levodopa values after broad bean (Vicia faba) consumption.
J Neurol Neurosurg Psychiatry. 55:725-727.



Rashid, KY and Bernier, CC 1993. Genetic diversity among isolates of Rhizoctonia solani and sources of resistance in Vicia faba.
Can J Plant Pathol. 15:23-28.

Reddy, MP, Sarla, N and Siddiq, EA 2002. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica. 128:9-17.
Rohlf, FJ 2000. Statistical power comparisons among alternative morphometric methods.
Am J Phys Anthropol. 111:463-478.


Rubio, J, Hajj-Moussa, E, Kharrat, M, Moreno, MT, Millan, T and Gill, J 2003. Two genes and linked RAPD markers involved in resistance to Fusarium oxysporum f. sp. ciceris race 0 in chickpea.
Plant Breed. 122:188-191.

Sahoo, MR, DasGupta, M, Kole, PC, Bhat, JS and Mukherjee, A 2007. Antioxidative enzymes and isozymes analysis of taro genotypes and their implications in Phytophthora blight disease resistance.
Mycopathologia. 163:241-248.


Saikia, R, Singh, BP, Kumar, R and Arora, DK 2005. Detection of pathogenesis-related proteins-chitinase and ╬▓-1,3-glucanase in induced chickpea. Curr Sci. 89:659-663.
Shen, SH, Chr├®tien, P, Bastien, L and Slilaty, SN 1991. Primary sequence of the glucanase gene from Oerskovia xanthineolytica. Expression and purification of the enzyme from Escherichia coli.
J Biol Chem. 266:1058-1063.


Singh, AK, Rai, VP, Chand, R, Singh, RP and Singh, MN 2013. Genetic diversity studies and identification of SSR markers associated with Fusarium wilt (Fusarium udum) resistance in cultivated pigeonpea (Cajanus cajan).
J Genet. 92:273-280.


Singh, M, Chaudhuri, I, Mandal, SK and Chaudhuri, RK 2011. Development of RAPD markers linked to Fusarium wilt resistance gene in castor bean (Ricinus communis L.). Genet Eng Biotechnol J. 28:1-9.
Snyder, WC and Hansen, HN 1940. The species concept in Fusarium.
Am J Bot. 27:64-67.

Souframanien, J and Gopalakrishna, T 2006. ISSR and SCAR markers linked to the mungbean yellow mosaic virus (MYMV) resistance gene in blackgram (Vigna mungo (L.) Hepper).
Plant Breed. 125:619-622.

Srivastava, RK, Mishra, SK, Singh, AK and Mohapatra, T 2012. Development of a coupling-phase SCAR marker linked to the powdery mildew resistance gene ŌĆśer1ŌĆÖ in pea (Pisum sativum L.).
Euphytica. 186:856-866.

Tanksley, SD, Young, ND, Paterson, AH and Bonierbale, MW 1989. RFLP mapping in plant breeding: new tools for old science.
Nat Biotechnol. 7:257-264.

Tekalign, A, Derera, J, Sibiya, J and Mumm, RH 2019. Molecular analysis for genetic diversity and population structure of Ethiopian faba bean (Vicia faba L.) accessions. Plant Biol Crop Res. 1:1010.
Terzopoulos, PJ and Bebeli, PJ 2008. Genetic diversity analysis of Mediterranean faba bean (Vicia faba L.) with ISSR markers.
Field Crops Res. 108:39-44.

Tufan, H and Erdo─¤an, C 2017. Genetic diversity in some faba bean (Vicia faba L.) genotypes assessed by simple sequence repeats.
Biotechnol Biotechnol Equip. 31:29-35.

Vandemark, GJ, Ariss, JJ, Bauchan, GA, Larsen, RC and Hughes, TJ 2006. Estimating genetic relationships among historical sources of alfalfa germplasm and selected cultivars with sequence related amplified polymorphisms.
Euphytica. 152:9-16.

Varshney, RK, Hoisington, DA and Tyagi, AK 2006. Advances in cereal genomics and applications in crop breeding.
Trends Biotechnol. 24:490-499.


Velasco-Ram├Łrez, AP, Torres-Mor├Īn, MI, Molina-Moret, S, S├Īnchez-Gonz├Īlez, JDJ and Santacruz-Ruvalcaba, F 2014. Efficiency of RAPD, ISSR, AFLP and ISTR markers for the detection of polymorphisms and genetic relationships in camote de cerro (Dioscorea spp.).
Electron J Biotechnol. 17:65-71.

Wang, Z, Wang, J-E, Wang, X-M, Gao, H-W, Dzyubenko, NI and Chapurin, VF 2012. Assessment of genetic diversity in Galega officinalis L using ISSR and SRAP markers.
Genet Resour Crop Evol. 59:865-873.

Yahia, Y, Hannachi, H and Ferchichi, A 2014. Genetic diversity of Vicia faba L. based on random amplified polymorphic DNA and simple sequence repeat markers.
Acta Bot Gallica Bot Lett. 161:151-158.

Yang, T, Bao, S-Y, Ford, R, Jia, T-J, Guan, J-P, He, Y-H, Sun, X-L, Jiang, J-Y, Hao, J-J, Zhang, X-Y and Zong, X-X 2012. High-throughput novel microsatellite marker of faba bean via next generation sequencing.
BMC Genomics. 13:602



Y├╝zba┼¤io─¤lu, E, A├¦ik, L and ├¢zcan, S 2008. Seed protein diversity among lentil cultivars.
Biol Plant. 52:126-128.

Zeid, M, Sch├Čn, C-C and Link, W 2003. Genetic diversity in recent elite faba bean lines using AFLP markers.
Theor Appl Genet. 107:1304-1314.


Zietkiewicz, E, Rafalski, A and Labuda, D 1994. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification.
Genomics. 20:176-183.






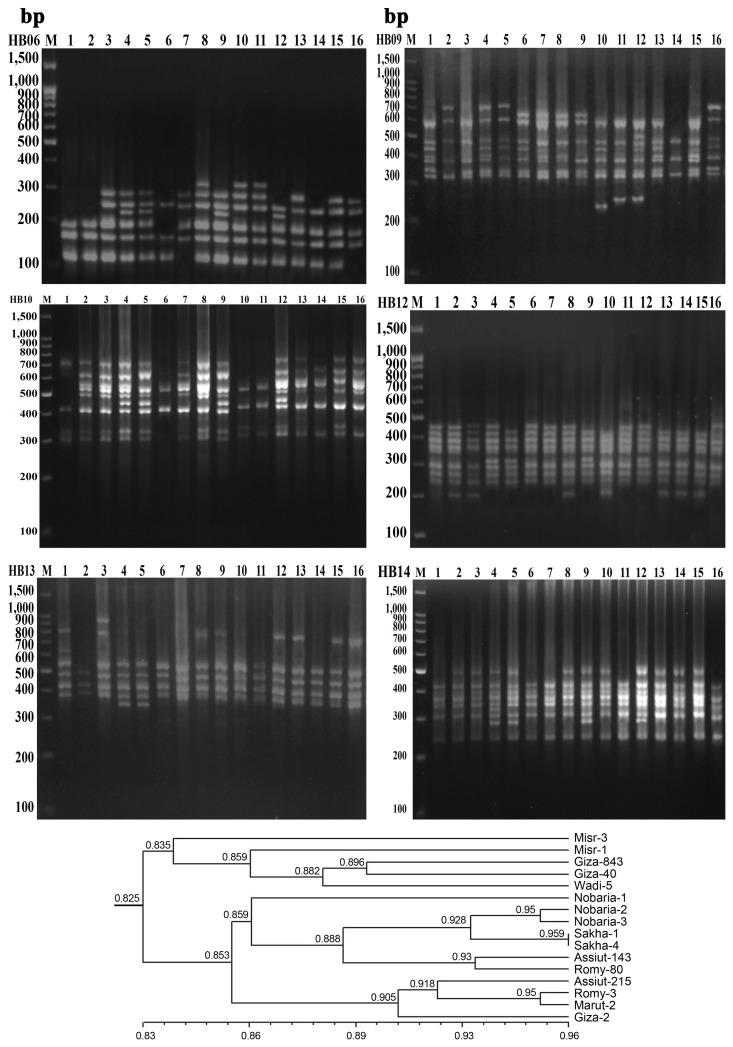
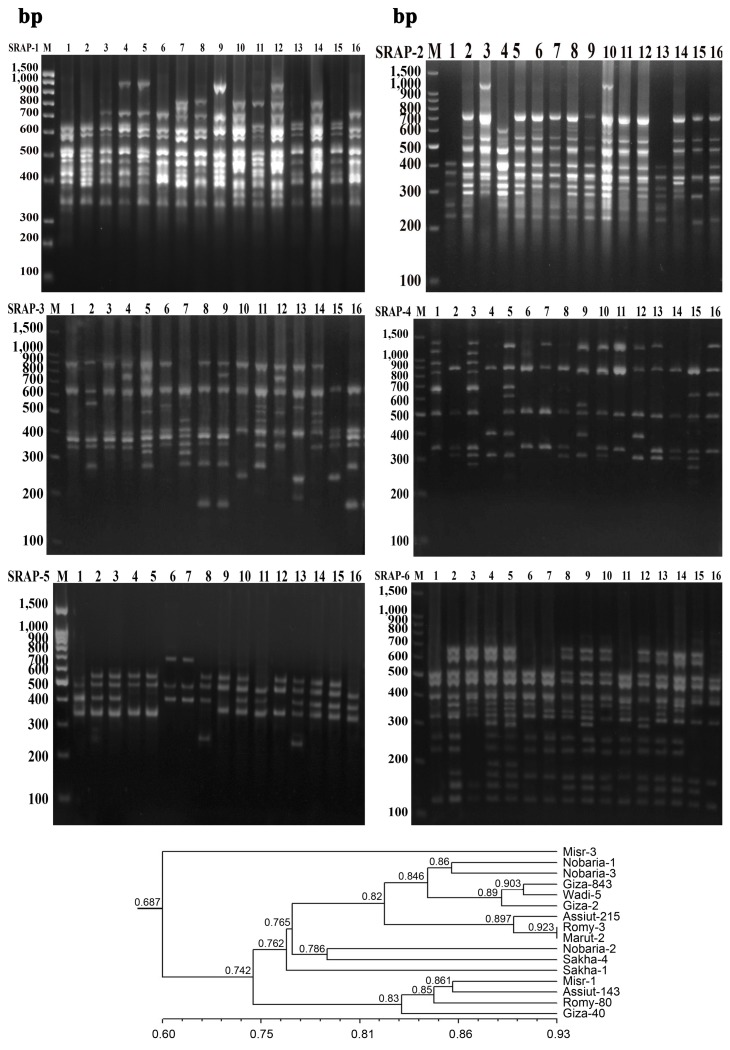
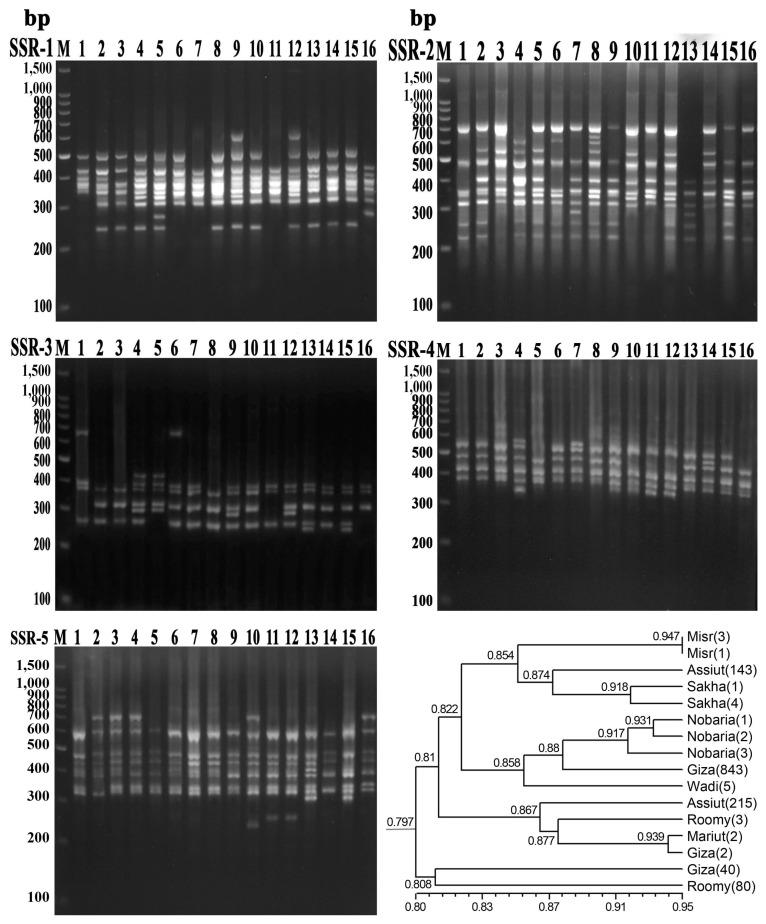
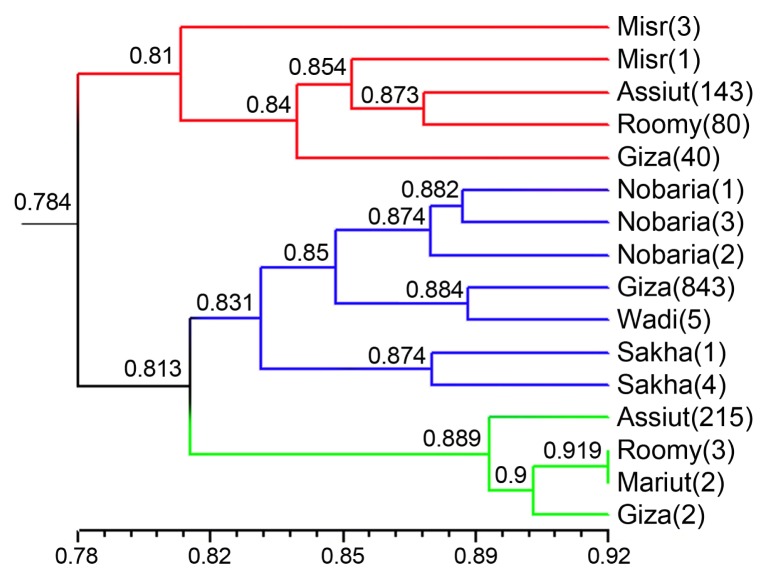
 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print






