 |
 |
| Plant Pathol J > Volume 35(4); 2019 > Article |
Abstract
For several years, temperatures in the Korean peninsula have gradually increased due to climate change, resulting in a changing environment for growth of crops and vegetables. An associated consequence is that emerging species of insect vector have caused increased viral transmission. In Jeju Island, Korea, occurrences of viral disease have increased. Here, we report characterization of five newly collected turnip mosaic virus (TuMV) isolates named KBJ1, KBJ2, KBJ3, KBJ4 and KBJ5 from a survey on Jeju Island in 2017. Full-length cDNAs of each isolate were cloned into the pJY vector downstream of cauliflower mosaic virus 35S and bacteriophage T7 RNA polymerase promoters. Their fulllength sequences share 98.9-99.9% nucleotide sequence identity and were most closely related to previously reported Korean TuMV isolates. All isolates belonged to the BR group and infected both Chinese cabbage and radish. Four isolates induced very mild symptoms in Nicotiana benthamiana but KBJ5 induced a hypersensitive response. Symptom differences may result from three amino acid differences uniquely present in KBJ5; Gly(382)Asp, Ile(891)Val, and Lys(2522)Glu in P1, P3, and NIb, respectively.
Turnip mosaic virus (TuMV) is a member of the genus Potyvirus within the family Potyviridae of plant viruses (Adams et al., 2011; Fauquet et al., 2005). Potyviruses have monopartite single-stranded positive-sense RNA genomes of 9,500-10,000 nt that are encapsulated in long flexuous filamentous particles of 700-750 nm in length. Because TuMV infects and damages a wide range of both crop and ornamental plants, including plants in the family Brassicaceae, it is an economically important virus. TuMV is distributed worldwide and has been reported from tropical regions of Africa, Asia, Oceania and North/South America (Provvidenti, 1996). Most TuMV isolates were collected from the family Brassicaceae, especially the genera Brassica and Raphanus but also several non-brassicaceous species (Ohshima et al., 2002).
TuMV is transmitted by a wide range of aphid species in a non-persistent manner. Climatic conditions that favor the build-up of aphid populations increase the spread of the virus. Consequently, climate change is altering the growing environment, affecting the production crops in the region, including Chinese cabbage, radish, rice, and soybean (Mckeown et al., 2004, Oh et al., 2015). Recently, incidences of insect-transmitting viruses have increased in China, Korea, and Japan (Yasaka et al., 2017). The weather patterns in East Asia are becoming monsoon-like, with heavy summer rainfall and severely cold winters (Ye et al., 2017). Therefore, the production of major crops and vegetables in these three countries are impacted by similar viral diseases. In addition to wind-assisted movement of insect vectors from west to east, increased agricultural trading between these countries enhances the movement of viral diseases (Tomimura et al., 2003).
Recent studies showed that isolates of TuMV and soybean mosaic virus (SMV) from Korea, both of which are insect- and seed-transmitted, were closely related to isolates from Japan and China, respectively (Han et al., 2016; Li et al., 2014). A previous report revealed Chinese and Japanese TuMV isolates originated from discrete lineages of the same population (Tomitaka and Ohshima, 2006).
TuMV isolates from around the world have been classified into four major groups based on either their biological phenotypes or phylogenetic relationships of their full genome sequences. At the biological phenotype level, isolates of the [B]-host type infect Brassica species systemically, inducing mosaic symptoms, but do not infect Raphanus (radish), while [(B)]-host type isolates occasionally infect Brassica latently, but also do not infect radish. Isolates of the [BR]-host type infect both Brassica and Raphanus, inducing systemic mosaic symptoms, and [B(R)]-host type isolates systemically infect Brassica with systemic mosaic and occasionally infect Raphanus. Phylogenetic analysis separates isolates into the basal-B (Brassica), world-B, basal-BR (Brassica/Raphanus), and Asian-BR groups. Among these groupings, the basal-B phylogenetic group, and [B]-host type group are the most variable, and isolates originate from diverse hosts including the Brassicaceae and a variety of other plant families (Ohshima et al., 2002; Tomimura et al., 2004; Tomitaka and Ohshima, 2006; Nguyen et al., 2013a, 2013b).
TuMV was recognized as a major cause of yield loss to radish and Chinese cabbage in the Gangwon area of South Korea (Choi and Choi, 1992; Hahm, 1995; Hahm et al., 1998). TuMV together with cucumber mosaic virus (CMV) and radish mosaic virus (RaMV) were also reported from a nationwide survey of radish fields conducted in South Korea in 2014 (Chung et al., 2015). Recently, Gong et al. (2019) collected new TuMV isolates from South Korea that showed symptoms of varying severity on Chinese cabbage, Nicotiana benthamiana and radish. Although new TuMV isolates were reported from Korea recently, a detailed genetic analysis has not been conducted of TuMV isolates from Jeju Island where 30% of Korea’s vegetables including carrots, Chinese cabbage, and radish are produced (Kim et al., 2012). Analysis of TuMV isolates from Jeju Island would provide information useful to study the lineage and movement of TuMV isolates among China, Japan and mainland Korea.
In our current study we constructed full-length infectious clones from TuMV isolates collected from Jeju Island in 2017. Our study focused on how closely the Jeju Island TuMV isolates are related to isolates from mainland Korea to determine the probable origin of the Jeju Island isolates, and their potential symptom determinants.
TuMV isolates from Jeju Island were collected in 2017 from radish fields in the Andeok region. The collected radish tissue showed mosaic and mottle symptoms. The virus infection was confirmed by RT-PCR using TuMV-specific primers (5′-TCT CAA TGG TTT AAT GGT CTG G-3′ and 5′-AAC CCC TTA ACG CCA AGT AAG-3′). From collected tissues we obtained 10 TuMV full-length cDNA constructs in the pJY binary vector as described by Han et al. (2016) and Park et al. (2017). All constructed clones were evaluated for pathogenesis using agroinfiltration of N. benthamiana as described by Lim et al. (2009). N. benthamiana plants were infiltrated with each of the clones and maintained in a growth chamber at 22-24°C or held in the lab for 24 h and then placed in the growth chamber. Seven of the full-length clones showed symptoms when inoculated on N. benthamiana. The full-length 9,833-nt sequences of seven infectious pJY::TuMV cDNA clones were determined by Macrogen Inc (Seoul, Korea). Sequences were assembled using CodonCode Aligner (CodonCode Corporation). Four clones showed unique sequence differences, and the other three clones were identical. Finally, we were named the five TuMV isolates as KBJ1, KBJ2. KBJ3, KBJ4 and KBJ5.
To evaluate the different pathogenicity of the five TuMV isolates, N. benthamiana plants were inoculated separately with each of the full-length clones by agroinfiltration. Mild mosaic and mottle symptoms appeared with each of the five isolates at 5-7 days post inoculation (dpi). At 7-14 dpi, the plants inoculated with isolates KBJ1-4 show severe leaf curling, mosaic and growth inhibition or partial necrosis. Plants inoculated with KBJ5 showed initial symptoms similar to the other isolates, but in addition developed stem necrosis. KBJ5 induced systemic necrosis at around 21 dpi, while isolates KBJ1-4 produced only mild symptoms with significant stunting (Fig. 1). Upper leaves of additional N. benthamiana plants inoculated with each isolate were harvested at 5 dpi and used to inoculate three 2-week-old seedlings each of R. sativus cvs. Chunghua and Iljin. At 14 dpi, no symptoms were observed with KBJ1, while varying degrees of mosaic were observed with the other four isolates (Fig. 1); all radish plants of each cv. were infected, as determined by RT-PCR.
For each sequence, the large open reading frame was translated using the Translate tool (http://web.expasy.org/translate/) to produce polyprotein sequences for each isolate of 3,164 amino acids. Complete predicted amino acid and nucleotide sequences were aligned separately using ClustalW. To investigate the region responsible for the systemic necrosis of KBJ5, we first compared the full-length sequences among these five isolates. The KBJ4 isolate share the highest identity with KBJ5, with 99.93% nucleotide (nt) sequence and 99.84% amino acid (aa) sequence identity. There are only five aa differences between the two isolates, residues 47 (P1 region), 382, 394 (HC-Pro region), 891 (P3 region), and 2522 (NIb region) (Fig. 2A). Isolates KBJ1-3 differed from KBJ5 by between 14 and 23 aa residues. Among all of the different aa residues, only three differences were shared by isolates KBJ1-4, which all induce mild symptoms in N. benthamiana. Interestingly, only isolate KBJ5 had Asp at aa 382 in P1, Val at aa 891 in P3, and Glu at aa 2522 in NIb, where the four mild isolates all had Gly, Ile, and Lys, respectively, at those positions. Potyvirus proteins, including HC-Pro, P3, CI and CP, have been identified as the virulence and avirulence determinants in various hosts (Gal-On, 2000; Jenner et al., 2002, 2003; Redondo et al., 2001; Sáenz et al., 2000, 2001; Suehiro et al., 2004). Through analysis of Jeju Island isolates we found aa variability in HC-Pro, P3, and NIb showed different symptom development, and could be potential candidates to analyze host and virus gene interactions.
Additional TuMV sequences were obtained from NCBI Genbank. Phylogenetic relationship of TuMV isolates in this work were analyzed together with twenty previously reported Korean TuMV isolates (Gong et al., 2019; Han et al., 2016) and 33 previously characterized TuMV isolates (Nguyen et al., 2013a) from NCBI. All isolates were compared based on aa and nt sequences. Phylogenetic trees were constructed by MEGA6 using the maximum-likelihood algorithm with 1000 bootstrap replicates (Hall, 2013). TuMV isolates were categorized into four previously described groups (Nguyen et al., 2013a, 2013b). The pathotypes of TuMV isolates were divided into two types: Brassica (B: can infect Brassica spp. but not Raphanus spp.) and Brassica-Raphanus (BR: can infect both Brassica and Raphanus spp.) (Ohshima et al., 2002). All of the Korean isolates examined fell within the Basal-BR group, with the Jeju isolates intermingled among the previously characterized Korean isolates. A further minor clade of the Basal-BR group is composed of Italian isolates ITA1 and ITA2, with Italian isolate Cal1 and Japanese isolate KWB779J falling between the other two Italian isolates and the large group of weakly separated Korean sequences including all five Jeju isolates (Fig. 3). There were close relationships between individual Jeju Island isolates and specific previously reported Korean isolates: Jeju isolate KBJ1 was grouped with DJ1, DJ5, DJ4; KBJ2 and KBJ3 were most closely related to DJ2; while KBJ4 and KJB5 grouped with isolate D6. Isolates DJ1-D6 were originally from a radish field in Daejeon, Korea. These results imply that Jeju Island TuMV isolates originated from the mainland of Korea rather than from neighboring countries.
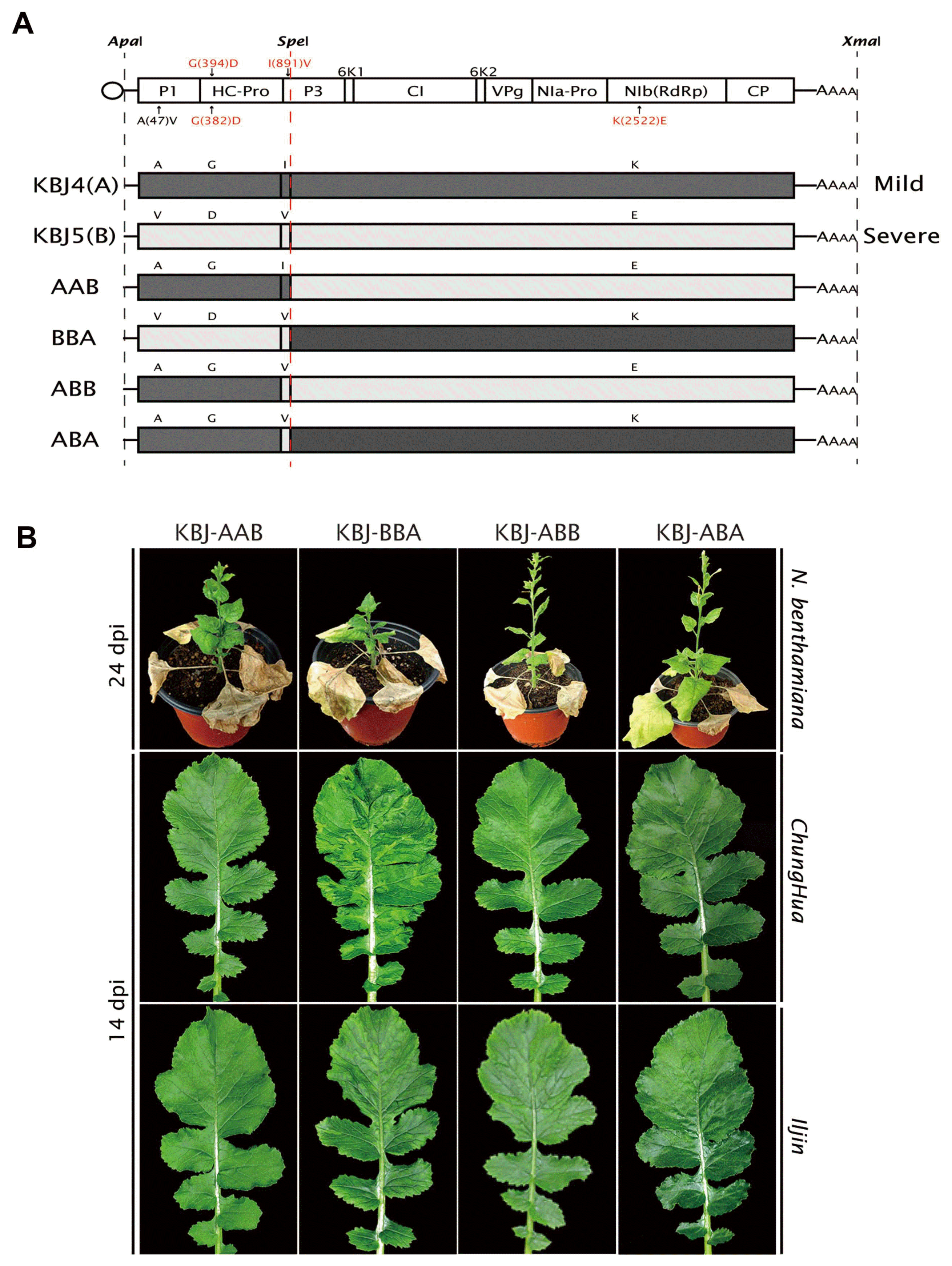
Here we report the complete genomic sequences of five newly developed TuMV infectious clones from Jeju Island, Korea. Interestingly four of the Jeju isolates produced relatively mild symptoms in N. benthamiana, whereas only KBJ5 induced stem necrosis and systemic foliar necrosis in N. benthamiana, and differed by only five amino acid residues from mild isolate KBJ4. As three of these differential residues were also distinct from KBJ1, KBJ2 and KBJ3, it was probable that one or more of these three residues (D382 in HC-Pro; V891 in P3; and E2522 in NIb) are singly or jointly determinant(s) of mild and severe necrosis. To address this, we generated chimeric constructs AAB and BBA between KBJ4 and KBJ5 by exchange of a 5′-terminal ApaI-SpeI (2829 bp) restriction fragment including the differential P1, HC-Pro, and P3 residues (Fig. 4A). The change I(891)V was also made in the KBJ4 ApaI-SpeI fragment by PCR substitution of a single nucleotide change in the SpeI-containing reverse primer (5′-AAACTAGT AAAACAGCTA CATTGCTGTC CGCTCTGACG-3′; SpeI site underlined, altered nt in bold), allowing creation of additional chimeric constructs ABB and ABA (Fig. 4A). Each of the chimeric constructs was then inoculated to both N. benthamiana and R. sativus cvs. Chunghua and Iljin, as before. Chimera BBA induced the most severe symptoms in both N. benthamiana (severe mosaic and growth inhibition) and the two radish cvs, (obvious mosaic), but without the systemic necrosis in N. benthamiana induced by KBJ5; chimeras AAB, ABB, and ABA all induced mild symptom more typical of KBJ4 in N. benthamiana and both radish cultivars (Fig. 4B). From these results it appears that both the KBJ5 N-terminal region containing differential P1, HC-Pro and P3 residues and the C-terminal portion including NIb (E2522) are required for the necrosis induced by KBJ5. However, comparison of BBA (severe) to ABA (mild) suggests that the region including P1 and HC-Pro residues derived from KBJ5 are important for symptom severity, while because AAB and KBJ4 induce the same symptoms, NIb (E2522) is not significant by itself; similarly, as ABA, ABB, and KBJ4 have the same symptoms, neither P3 (V891) alone, or P3 (V891) + NIb (E2522) are sufficient to induce severe symptoms. As HC-Pro (D382) is the only other residue unique to KBJ5, this residue is presumable required for severe symptoms, in combination with NIb (E2522) and possibly P3 (V891).
It is not unusual for potyvirus symptoms in different hosts or even cultivars to depend on differential residues in distinct gene products including P3 (Jenner et al., 2002, 2003; Saenz et al., 2000) and other gene products (Salvador et al., 2008), and we have previously identified apparent interactions between determinants in two TuMV gene products affecting host reactions (Kim et al., 2019). However, in the current case, symptom severity co-varied in N. benthamiana and R. sativus, and the influence of P3 (V891) may either increase or decrease symptom severity dependent on the context of other differential residues.
In addition to the identification of residues potentially associated with symptom severity in N. benthamiana and radish, the first full length sequences of TuMV isolates in Jeju Island reported here will contribute to understanding the potential relationships among Eastern Asian TuMV isolates.
Acknowledgments
This work was supported by a grant from the Next-Generation BioGreen 21 Program (Project No. PJ01365501), Rural Development Administration, Republic of Korea.
Fig. 1
Symptoms of the five isolates (KBJ1-KBJ5) inoculated to Nicotiana benthamiana, shown at 7 days post inoculation (dpi) and 21 dpi; and to radish (Raphanus sativus) cultivars Chunghua and Iljin at 14 dpi. Representative plants inoculated with each isolate are shown. TuMV infection of all radish plants was confirmed by RT-PCR.
Fig. 2
Amino acid residues differing among isolates KBJ1-5. Isolate KBJ-5 is marked in red because it can induce systemic necrosis in Nicotiana benthamiana. Differing amino acid residues are shown in red or blue, with unique KBJ5 residues 382 (HC-Pro), 891 (P3) and 2522 (NIb) marked by a red box.
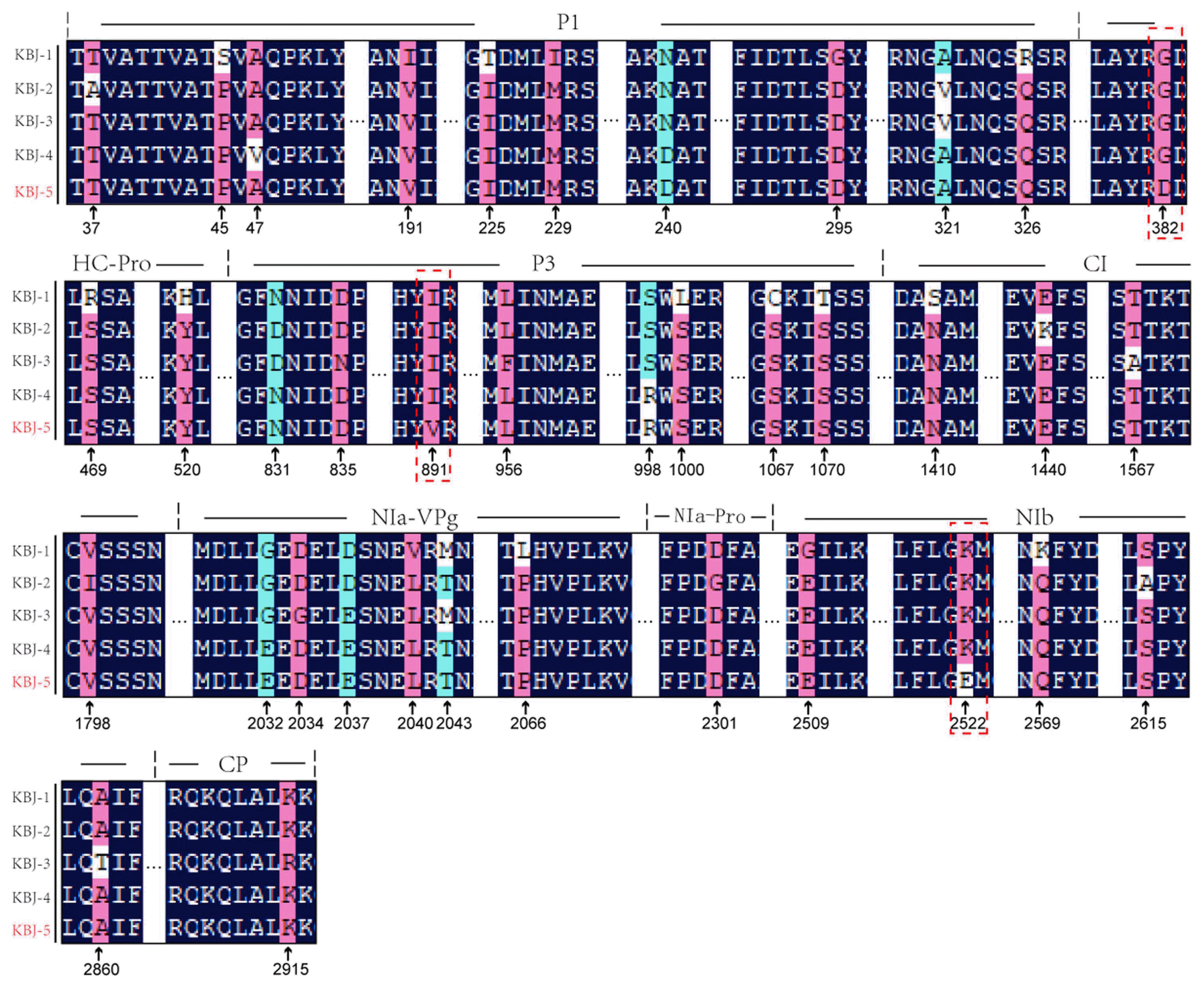
Fig. 3
Phylogenetic tree constructed by the maximum-likelihood method with 1,000 bootstrap replicates, based on the complete genome sequences of 58 TuMV isolates including twenty other Korean isolates (Gong et al., 2019) and the five new isolates from Jeju (indicated by arrows to the right of the isolate/accession number/country label).
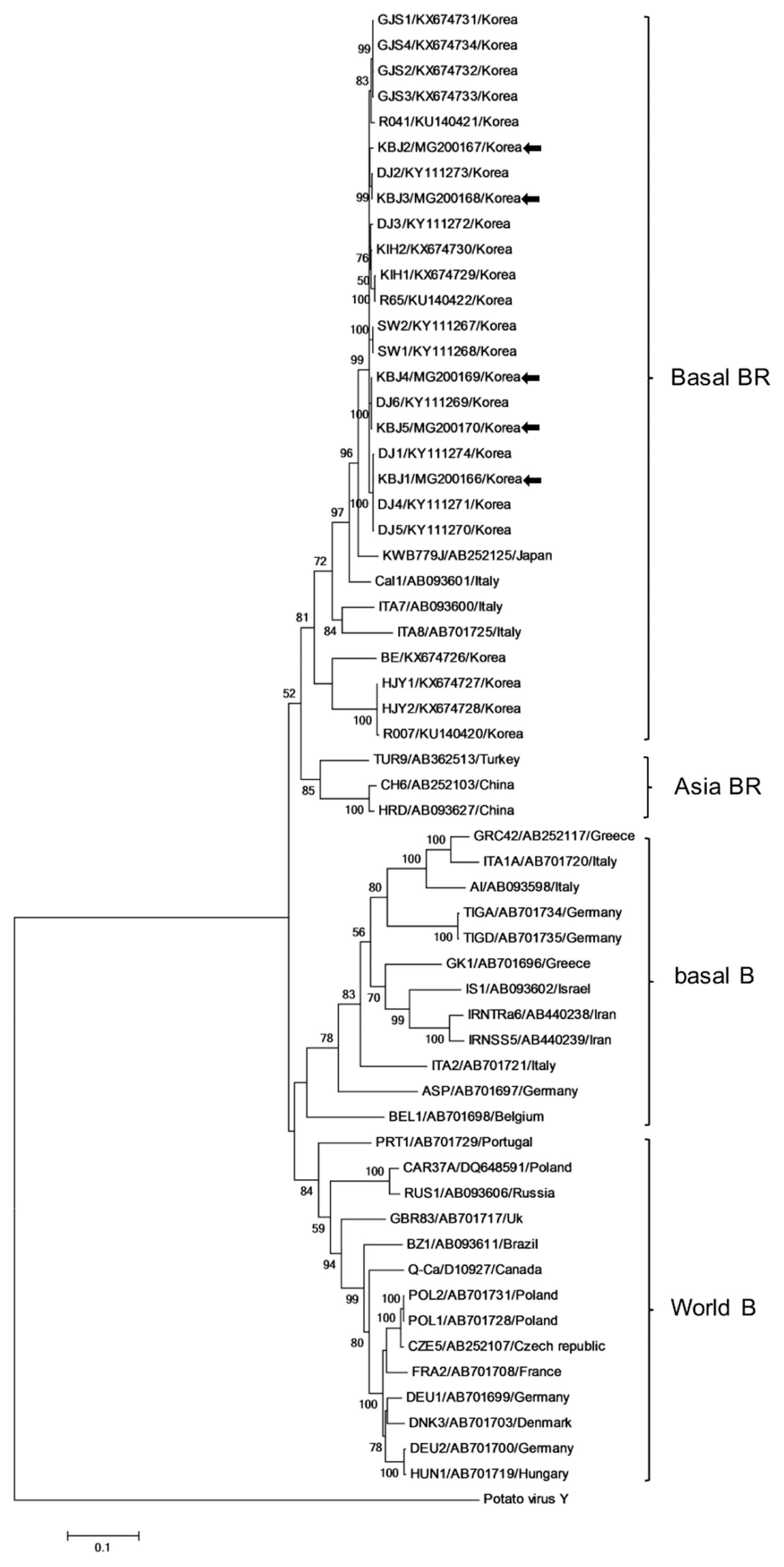
Fig. 4
(A) Structure of the chimeras constructed between mild isolate KBJ4 and severe isolate KBJ5. Chimeras AAB and BBA were generated by exchange of a 5′ ApaI/SpeI fragment; to create chimeras ABB and ABA, mutagenic PCR was utilized to alter a single nt resulting in I891V in the ApaI/SpeI fragment. (B) Each chimeric construct was then used to infect Nicotiana benthamiana, and radish cultivars Chunghua and Iljin. Images of representative plants were captured at 24 days post inoculation (dpi) for N. benthamiana, and 14 dpi for radish.
References
Adams, MJ, Zerbini, FM, French, R, Rabenstein, F, Stenger, DC and Valkonen, JPT 2011. Potyviridae. In: Virus taxonomy: Ninth report of the international committee on taxonomy of viruses, eds. by AMQ King, MJ Adams, EB Carstens and EJ Lefkowitz, 1069-1089. Elsevier Academic Press, San Diego, CA, USA.

Chung, JS, Han, JY, Kim, JK, Ju, HK, Gong, JS, Seo, EY, Hammond, J and Lim, HS 2015 Survey of viruses present in radish fields in 2014. Res Plant Dis. 21:235-242 (in Korean).

Fauquet, CM, Mayo, MA, Maniloff, J, Desselberger, U and Ball, LA 2005. Virus taxonomy: Eighth report of the international committee on taxonomy of viruses. Elsevier Academic Press, San Diego, CA, USA. 1162.
Gal-On, A 2000. A point mutation in the FRNK motif of the potyvirus helper component-protease gene alters symptom expression in cucurbits and elicits protection against the severe homologous virus. Phytopathology. 90:467-473.


Gong, J, Ju, HK, Kim, I-H, Seo, E-Y, Cho, I-S, Hu, W-X, Han, J-Y, Kim, J-K, Choi, SR, Lim, YP, Hammond, J and Lim, H-S 2019. Sequence variations among seventeen new radish isolates of Turnip mosaic virus showing differential pathogenicity and infectivity in Nicotiana benthamiana, Brassica rapa, and Raphanus sativus. Phytopathology. 109:904-912.


Hall, BG 2013. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 30:1229-1235.


Hahm, YI 1995 Recent occurrence of TuMV disease on radish and Chinese cabbage in alpine region, Kangwon province. Res Plant Dis. 1:45-46 (in Korean)..
Hahm, YI, Kwon, M, Kim, JS, Seo, HY and Ahn, JH 1998 Surveys on disease occurrence in major horticultural crops in Kangwon alpine areas. Korean J Plant Pathol. 14:668-675 (in Korean)..
Han, JY, Chung, J, Kim, J, Seo, EY, Kilcrease, JP, Bauchan, GR, Lim, S, Hammond, J and Lim, HS 2016. Comparison of helper component-protease RNA silencing suppression activity, subcellular localization, and aggregation of three Korean isolates of Turnip mosaic virus. Virus Genes. 52:592-596.


Jenner, CE, Tomimura, K, Ohshima, K, Hughes, SL and Walsh, JA 2002. Mutations in Turnip mosaic virus P3 and cylindrical inclusion proteins are separately required to overcome two Brassica napus resistance genes. Virology. 300:50-59.


Jenner, CE, Wang, X, Tomimura, K, Ohshima, K, Ponz, F and Walsh, JA 2003. The dual role of the potyvirus P3 protein of Turnip mosaic virus as a symptom and avirulence determinant in brassicas. Mol Plant-Microbe Interact. 16:777-784.


Kim, JS, Lee, SH, Choi, HS, Kim, MK, Kwak, HR, Kim, JS, Nam, M, Cho, JD, Cho, IS and Choi, GS 2012. 2007-2011 characteristics of plant virus infections on crop samples submitted from agricultural places. Res Plant Dis. 18:277-289.

Kim, I-H, Ju, H-K, Gong, J, Han, J-Y, Seo, E-Y, Cho, S-W, Hu, W-X, Choi, SR, Lim, YP, Domier, L, Hammond, J and Lim, H-S 2019 A Turnip mosaic virus determinant of systemic necrosis in Nicotiana benthamiana, and a novel resistance-breaking determinant in Chinese cabbage identified from chimeric infectious clones. Phytopathology. (in press).


Lim, HS, Bragg, JN, Ganesan, U, Ruzin, S, Schichnes, D, Lee, MY, Vaira, AM, Ryu, KH, Hammond, J and Jackson, AO 2009. Subcellular localization of the Barley stripe mosaic virus triple gene block proteins. J Virol. 83:9432-9448.



Li, MJ, Kim, JK, Seo, EY, Hong, SM, Hwang, EI, Moon, JK, Domier, LL, Hammond, J, Youn, YN and Lim, HS 2014. Sequence variability in the HC-Pro coding regions of Korean Soybean mosaic virus isolates is associated with differences in RNA silencing suppression. Arch Virol. 159:1373-1383.


Mckeown, AW, Warland, J, Mcdonald, MR and Hutchinson, CM 2004. Cool season crop production trends: A possible signal for global warming. Acta Hortic. 638:241-248.

Nguyen, HD, Tran, HT and Ohshima, K 2013a. Genetic variation of the Turnip mosaic virus population of Vietnam: a case study of founder, regional and local influences. Virus Res. 171:138-149.


Nguyen, HD, Tomitaka, Y, Ho, SYW, Duchêne, S, Vetten, HJ, Lesemann, D, Walsh, JA, Gibbs, AJ and Ohshima, K 2013b. Turnip mosaic potyvirus probably first spread to Eurasian brassica crops from wild orchids about 1000 years ago. PLoS One. 8:e55336


Ohshima, K, Yamaguchi, Y, Hirota, R, Hamamoto, T, Tomimura, K, Tan, Z, Sano, T, Azuhata, F, Walsh, JA, Fletcher, J, Chen, J, Gera, A and Gibbs, A 2002. Molecular evolution of Turnip mosaic virus: evidence of host adaptation, genetic recombination and geographical spread. J Gen Virol. 83:1511-1521.


Oh, S, Moon, KH, Song, EY, Son, IC and Koh, SC 2015. Photosynthesis of Chinese cabbage and radish in response to rising leaf temperature during spring. Hortic Environ Biotechnol. 56:159-166.

Park, CH, Ju, HK, Han, JY, Park, JS, Kim, IH, Seo, EY, Kim, JK, Hammond, J and Lim, HS 2017. Complete nucleotide sequences and construction of full-length infectious cDNA clones of Cucumber green mottle mosaic virus (CGMMV) in a versatile newly developed binary vector including both 35S and T7 promoters. Virus Genes. 53:286-299.


Provvidenti, R 1996. Turnip mosaic potyvirus. In: Viruses of plants, eds. by AA Brunt, K Crabtree, MJ Dallwitz, AJ Gibbs and L Watson, 1340-1343. CAB International, Wallingford, UK.
Redondo, E, Krause-Sakate, R, Yang, SJ, Lot, H, Le Gall, O and Candresse, T 2001. Lettuce mosaic virus pathogenicity determinants in susceptible and tolerant lettuce cultivars map to different regions of the viral genome. Mol Plant-Microbe Interact. 14:804-810.


Sáenz, P, Cervera, MT, Dallot, S, Quiot, L, Quiot, JB, Riechmann, JL and García, JA 2000. Identification of a pathogenicity determinant of plum pox virus in the sequence encoding the C-terminal region of protein P3+6K(1). J Gen Virol. 81:557-566.


Sáenz, P, Quiot, L, Quiot, JB, Candresse, T and García, JA 2001. Pathogenicity determinants in the complex virus population of a Plum poxvirus isolate. Mol Plant-Microbe Interact. 14:278-287.


Suehiro, N, Natsuaki, T, Watanabe, T and Okuda, S 2004. An important determinant of the ability of Turnip mosaic virus to infect Brassica spp. and/or Raphanus sativus is in its P3 protein. J Gen Virol. 85:2087-2098.


Tomimura, K, Gibbs, AJ, Jenner, CE, Walsh, JA and Ohshima, K 2003. The phylogeny of Turnip mosaic virus; comparisons of 38 genomic sequences reveal a Eurasian origin and a recent ‘emergence’ in East Asia. Mol Ecol. 12:2099-2111.


Tomimura, K, Spak, J, Katis, N, Jenner, CE, Walsh, JA, Gibbs, AJ and Ohshima, K 2004. Comparisons of the genetic structure of populations of Turnip mosaic virus in west and east Eurasia. Virology. 330:408-423.


Tomitaka, Y and Ohshima, K 2006. A phylogeographical study of the Turnip mosaic virus population in east Asia reveals an ‘emergent’ lineage in Japan. Mol Ecol. 15:4437-4457.



Yasaka, R, Fukagawa, H, Ikematsu, M, Soda, H, Korkmaz, S, Golnaraghi, A, Katis, N, Ho, SYW, Gibbs, AJ and Ohshima, K 2017. The timescale of emergence and spread of Turnip mosaic potyvirus. Sci Rep. 7:4240



Ye, J, Gong, Y and Fang, R 2017 Research progress and perspective of tripartite interaction of virus-vector-plant in vector-borne viral diseases. Bull Chin Acad Sci. 32:845-855 (in Chinese).
- TOOLS



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




