 |
 |
| Plant Pathol J > Volume 35(6); 2019 > Article |
Abstract
Like many fungal pathogens, the conidium and appressorium play key roles during polycyclic dissemination and infection of Magnaporthe oryzae. Ran-binding protein microtubule-organizing center (RanBPM) is a highly conserved nucleocytoplasmic protein. In animalia, RanBPM has been implicated in apoptosis, cell morphology, and transcription. However, the functional roles of RanBPM, encoded by MGG_00753 (named MoRBP9) in M. oryzae, have not been elucidated. Here, the deletion mutant ΔMorbp9 for MoRBP9 was generated via homologous recombination to investigate the functions of this gene. The ΔMorbp9 exhibited normal conidial germination and vegetative growth but dramatically reduced conidiation compared with the wild type, suggesting that MoRBP9 is involved in conidial production. ΔMorbp9 conidia failed to produce appressoria on hydrophobic surfaces, whereas ΔMorbp9 still developed aberrantly shaped appressorium-like structures at hyphal tips on the same surface, suggesting that MoRBP9 is involved in the morphology of appressorium-like structures from hyphal tips and is critical for development of appressorium from germ tubes. Taken together, our results indicated that MoRBP9 played a pleiotropic role in polycyclic dissemination and infection-related morphogenesis of M. oryzae.
The ascomycete fungal pathogen Magnaporthe oryzae causes rice blast disease, seriously decreasing global rice production (Talbot, 2003). Infection starts when M. oryzae conidia attach to rice leaves and sense surface signals (Zhou et al., 2009). Subsequently, a germ tube emerges and develops into a dome-shaped infection structure called an appressorium, which is capable of penetrating the rice cuticle (Yi et al., 2008). After successful invasion and colonization, asexually reproduced conidia disseminate to adjacent uninfected rice plants by wind and rain splash, initiating a new infection cycle (Odenbach et al., 2007). Both production of massive conidia and formation of functional appressoria therefore play indispensable roles in the polycyclic processes of M. oryzae.
Conidiogenesis and appressorium formation are involved in a cascade of morphological processes, which have remained difficult to decipher (Kim et al., 2009). Investigation of the related genetic mechanisms contributes to understanding those complex events. Previous studies have identified functional genes that regulate conidiation in M. oryzae. Mutation of MoHOX2/HTF1, COS1, CON6, and FLB4 genes completely abolished conidiation, suggesting their critical roles in conidiation (Kim et al., 2009; Liu et al., 2010; Matheis et al., 2017; Shi and Leung, 1995; Zhou et al., 2009). Targeted deletions of MGG1, MNH6, and MoCDC42 led to reduced conidiation, indicating that these genes are related to this process (Li et al., 2015; Lu et al., 2007; Zheng et al., 2009), and COM1 and MoSWI6 were found to regulate conidial morphology (Qi et al., 2012; Yang et al., 2010). A well-developed appressorium plays an indispensable role in M. oryzae preinfection of the host. Among the genetic regulatory mechanisms of appressorium development, signaling pathways orchestrate appressorium-mediated infection. The conserved mitogen-activated protein kinase transduction pathway controls appressorium development and subsequent hyphal infection inside plant cells (Jiang et al., 2018). PMK1 orthologous to yeast Fus3/ Kss1 regulates appressorium formation and plant cell-to-cell infection by M. oryzae (Jiang et al., 2018; Sakulkoo et al., 2018). The surface recognition and pathogenicity of M. oryzae are regulated by the cAMP-dependent protein kinase A (PKA) signaling pathway (Li et al., 2017). Appressorium maturation requires translocation and metabolism of lipids and glycogen, which are dependent on PMK1 and PKA (Thines et al., 2000). It is therefore of interest to investigate new genes involved in both conidiation and appressorium development.
Ran-binding protein microtubule-organization center (RanBPM or RanBP9), a ubiquitous protein localized in both the nucleus and cytoplasm, was initially found using a yeast two-hybrid system with the small GTPase Ran as bait (Nakamura et al., 1998; Salemi et al., 2017). The designated RanBPM was found to interact with nucleated microtubules at centrosomes (Nakamura et al., 1998). Moreover, this protein was also identified as the ninth member of the Ran-binding protein family, resulting in the name RanBP9 (Das et al., 2018). Further studies revealed that RanBPM does not have a Ran-binding domain and is incapable of binding to Ran GTPase in vivo (Das et al., 2018; Nishitani et al., 2001). Instead of a Ran-binding domain, RanBPM contains a spore lysis A and ryanodine receptor (SPRY) domain, as well as a lissencephaly type-I-like homology (LisH) motif, a carboxyl terminus with a LisH (CTLH) motif, and a CT11-RanBPM (CRA) domain from the N- to C-termini of the amino acid sequence (Tomaštíková et al., 2012; Wang et al., 2004). The SPRY and CRA domains have been suggested to contribute to RNA metabolism signaling and protein degradation, respectively (Das et al., 2018; Perfetto et al., 2013). The LisH and CTLH motifs have been shown to be involved in chromosome segregation and microtubule dynamics (Das et al., 2018). RanBP9 contains four conserved protein-protein interaction domains and is thought to interact with many protein partners and to integrate different signaling pathways in mammalian cells (Salemi et al., 2017). However, the functions of RanBP9 in fungi are still poorly understood.
In the present study, we aimed to elucidate the functions of MoRBP9 in M. oryzae by using a targeted gene deletion method. Our results showed that deletion of MoRBP9 did not influence vegetative growth or conidial germination but dramatically reduced conidiation and completely abolished appressorium formation on the hydrophobic surface. Exogenous cAMP only partly restored the abnormal development of the appressoria. Taken together, the results showed that MoRBP9 plays an important role in dissemination and preinfection development of M. oryzae.
The transformants were generated from M. oryzae wild-type strain KJ201. The fungal strains were grown under standard conditions using V8 agar (V8; 80 ml/l V8 juice and 15 g/l agar powder) or oatmeal agar (OMA; 50 g/l oatmeal and 15 g/l agar powder) at 25°C with constant fluorescent light. Mycelia grown in liquid CM (10 g/l sucrose, 6 g/l casamino acid, and 6 g/l yeast extract) were prepared for DNA and RNA extraction.
All sequences were obtained from online databases of the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/) and the Comparative Fungal Genomics Platform (http://cfgp.riceblast.snu.ac.kr/). Homologous sequences were searched using NCBI BLASTP (https://blast.ncbi.nlm.nih.gov/). Domain and motif structures were predicted using InterPro-Scan (https://www.ebi.ac.uk/interpro/). Conserved amino acid sequences were aligned using MEGA7 software and then edited using BioEdit, version 7.0.5 software (http://www.mbio.ncsu.edu/BioEdit/page2.html). Phylogenetic relationships were analyzed by the neighbor-joining method, with a bootstrap of 1,000, using MEGA7 software. All primers were synthesized by Bioneer (https://www.bioneer.co.kr/).
Total RNA was isolated from mycelia, mycelia bearing conidia, conidia, germinated conidia, and appressoria and infectious hyphae in rice leaves using Easy-Spin (iNtRON Biotechnology, Seongnam, Korea). The synthesis of first-strand complementary DNA (cDNA) from total RNA was performed using the SuperScript III First-strand Synthesis System (Invitrogen, Carlsbad, CA, USA). Reverse transcription polymerase chain reaction (RT-PCR) to detect transcripts of MoRBP9 in the transformants was conducted in a 20 μl mixture containing 25 ng cDNA, 20 U Pfu Plus DNA polymerase (Elpis Bio, Daejeon, Korea) and 10 pmol forward and reverse primers; the β-tubulin gene (MGG_00604) was used as a control (Supplementary Table 1). PCR was conducted on the StepOne Real-Time PCR System (Applied Biosystems, Foster, CA, USA) using the following program: 3 min at 95°C (1 cycle), followed by 20 s at 95°C, 30 s at 58°C, and 30 s at 72°C (30 cycles). Quantitative RT-PCR (qRT-PCR) was performed to analyze the expression levels of MoRBP9 during developmental stages as described previously (Han et al., 2018). The qRT-PCR mixture (10 μl) contained 5 μl HiPi Real-Time PCR 2× Master Mix (Elpis Bio), 1 μl cDNA (25 ng/μl), and 0.5 μl each primer (10 pmol/μl). PCR was conducted as follows: 3 min at 95°C (1 cycle), followed by 20 s at 95°C, 30 s at 58°C, and 30 s at 72°C (40 cycles). After normalizing the resulting cycle threshold (Ct) values to that of β-tubulin, relative gene expression was calculated as 2− ΔCt , where −ΔCt = (Ct, target gene − Ct, β-tubulin). The fold change in expression during different developmental stages was calculated as 2− ΔΔCt , where −ΔΔCt = (Ct, target gene − Ct, β-tubulin)test condition − (Ct, target gene − Ct, β-tubulin)mycelia (Han et al., 2015; Livak and Schmittgen, 2001). Three independent qRT-PCR experiments, with two replicates per experiment, were conducted.
Fungal genomic DNA used for PCR and Southern blot hybridization was prepared using a standard method (Han et al., 2015). The genomic DNA was digested with the restriction enzyme Hind III, separated via a 1% agarose gel, and transferred to Hybond N+ membranes (Amersham Pharmacia Biotech, Little Chalfont, UK). The DNA hybridization probe (approximately 500 bp) was amplified using the primers PF/PR (Supplementary Table 1) and labelled using Biotin-High Prime (Roche, Indianapolis, IN, USA). The probe hybridized to the membrane was detected using the ChemiDoc XRS + system and visualized using Image Lab software (Bio-Rad Laboratories, Hercules, CA, USA). Genomic DNA for screening experiments was isolated using a quick and safe DNA extraction method described previously (Chi et al., 2009).
Based on the M. oryzae genome, 1.5 kb sequences upstream and downstream of MoRBP9 were amplified using the primers 5F/5R and 3F/3R (Supplementary Table 1), respectively. In addition, the HPH cassette (1.5 kb) containing the hygromycin phosphotransferase gene was amplified using the primers HPHF/HPHR (Supplementary Table 1). The three amplicons were fused using double-joint PCR with the 5F/3R primers, and the resulting deletion constructs were amplified using the primers NF/NR (Supplementary Table 1) (Leung et al., 1990). Amplified deletion constructs were directly introduced into wild-type protoplasts using the polyethylene glycol method (Han et al., 2018; Leung et al., 1990). Transformants were selected by PCR-based screening using the primers SF/SR (Supplementary Table 1), and the putative targeted-gene-deletion mutants were subsequently purified using single spore isolation. Southern blotting and RT-PCR were performed to verify the correct deletion mutants. For ΔMorbp9 complementation, targeted genes in the wild-type genome were amplified using the cmF/cmR primers (Supplementary Table 1). The ΔMorbp9 protoplasts were transformed with amplicons of the targeted genes along with the pII99 vector containing the geneticin resistance gene. Complemented strains were screened by phenotypic restoration of ΔMorbp9 defects and were finally confirmed by RT-PCR.
For comparing vegetative growth, mycelial colony diameter was measured after growing the wild type and transformants on V8, oatmeal, and CM agar medium at 25°C without light for 6 days. The hydrophobicity of vegetative hyphae was tested by dropping sodium dodecyl sulfate (SDS) and water on 7-day-old mycelia. Conidiation was evaluated by counting conidia harvested in 5 ml distilled water from 7-day-old OMA medium. Lactophenol blue solution (Sigma-Aldrich, St. Louis, MO, USA) was used to distinguish conidiophores from mycelia (Kim et al., 2009). Conidial germination was performed by placing the conidial suspension (5 × 104 /ml) on hydrophobic coverslips and counting germinated conidia 2 h later. Appressorium formation was conducted by incubating a conidial suspension on hydrophobic coverslips and hydrophilic slide glasses with or without the addition of 5 mM exogenous cAMP (Fu et al., 2018). Surface hydrophobicity was tested by dropping water or SDS solution onto 14-day-old mycelia bearing conidia and evaluated by comparing mycelial wettability. Experiments were repeated three times with three replicates for each repeat. All data were analyzed according to Duncan’s test at P < 0.05.
Rice seedlings (at three- to four-leaf stage) of the susceptible rice cultivar Oryza sativa cv. Nakdongbyeo were sprayed with a conidial suspension (105/ml) containing Tween 20 (250 ppm) and incubated in a chamber at 25°C for 7 days. Wounded and unwounded leaf assays were conducted by inoculating a conidial suspension (105/ml) and mycelial agar plugs into wounded and unwounded rice leaves and then placing in humid boxes for 6 days. The appressorium penetration assay was performed using onion epidermis and rice sheath as described previously (Odenbach et al., 2007; Park et al., 2014). The experiments were repeated three times with three replicates for each repeat.
The MoRBP9 gene is predicted to encode a protein (759 amino acids) containing two conserved domains and two conserved motifs. To determine the relationship between MoRBP9 and its homologous proteins, we performed phylogenetic analyses, schematic representation of the domains, and alignments of the amino acids within the domains of MoRBP9 proteins from 10 different organisms belonging to fungi, animalia, and planta. The phylogenetic analyses showed that all proteins can be divided into three clades. MoRBP9 was closely related to orthologs from Fusarium graminearum, Botrytis cinerea, Colletotrichum higginsianum, and C. gloeosporioides but relatively distant to homologs from animalia and planta. Domain prediction indicated that all proteins had two domains and two motifs, except for the MoRBP9 homolog from O. sativa, which lacked the LIS1 motif. The domains and motifs are found to be closely present to C-termini of MoRBP9 homologues from fungi, compared with those from animalia and planta (Fig. 1A). Particularly, the CTLH motif and CRA domain of MoRBP9 orthologs from Arabidopsis thaliana and O. sativa shared five amino acids in the C- and N-termini, respectively (Fig. 1B). A NCBI BLASTP search showed that MoRBP9 shared 52% sequence identity with EQB53806.1 from C. gloeosporioides and 50% sequence identity with XP_018156055.1, XP_011309041.1, and XP_024552885.1 from C. higginsianum, F. graminearum, and B. cinerea, respectively. However, MoRBP9 only had 29% identity with NP_973963 from A. thaliana and 25% identity with XP_015627237.1 from O. sativa (Fig. 1B). These results indicate that MoRBP9 orthologs are well conserved in fungi.
To predict the function of MoRBP9 in M. oryzae development and pathogenicity, gene expression was evaluated at different stages of fungal growth by qRT-PCR (Fig. 2A). MoRBP9 was downregulated in conidia and germinated conidia, in which it showed induced expression in appressoria and invasive hyphae (Fig. 2A). Particularly, expression of MoRBP9 was strongly induced (~9.5-fold) in appressoria, suggesting that MoRBP9 is possibly involved in appressorium development. To determine the functional roles of MoRBP9, the homologous replacement method was used to generate a targeted-gene-deletion transformant (Fig. 2B). The transformants were selected by PCR analysis, and the mutants were verified by Southern blotting and RT-PCR (Fig. 2C and D). Genomic copies of MoRBP9 were reintroduced into ΔMorbp9 to generate a complementary transformant, Morbp9c, which was used to confirm ΔMorbp9 phenotypes resulting from targeted gene deletion. The sequence within the MoRBP9 gene was amplified by RT-PCR, and as expected, Morbp9c showed recovery of targeted gene expression (Fig. 2D).
To investigate the roles of MoRBP9 in fungal growth and development, we compared phenotypes between the wild type and targetedgene-deletion mutants. ΔMorbp9 exhibited no difference in the vegetative growth rate between complete and limited nutrition conditions but was relatively less pigmentated under both light and dark culture conditions. We then evaluated asexual reproduction and found that ΔMorbp9 produced approximately 2.4% of the amount of conidia compared with wild type, suggesting a dramatic impairment in conidiation (Fig. 3). However, this defect in asexual production was completely restored to wild-type levels in Morbp9c. To better evaluate the function of MoRBP9 in conidiation, we used a microscope to observe conidiophore and conidia during conidiation. The wild type was found to form a dense cluster of conidiophores bearing conidia, whereas ΔMorbp9 only produced very few conidia (Fig. 3A). To identify potential reasons for this phenotype, we used lactophenol blue staining to distinguish conidiophores from aerial hyphae. The results showed that conidiophores were rarely formed in ΔMorbp9, compared with many conidiophores observed in the wild type and Morbp9c, suggesting that MoRBP9 may regulate conidiation by controlling conidiophore formation (Fig. 3C). In M. oryzae, several genes such as HOX2, COS1, FLB3, FLB4, and CON6 were reported to regulate the conidiation process (Kim et al., 2009; Matheis et al., 2017; Shi and Leung, 1995; Zhou et al., 2009). To determine whether deletion of MoRBP9 affects expression of these conidiation-regulating genes, we performed qRT-PCR to measure their expression. The expression levels of COS1, FLB3, and FLB4 were greatly reduced in ΔMorbp9, compared with the wild type, indicating that MoRBP9 may modulate the expression of these conidiation-related genes (Fig. 3D). Overall, these results indicate that MoRBP9 plays a role in the early stage of conidiation.
As appressorium development is crucial for preinfection of M. oryzae, we performed appressorium formation assays on inductive hydrophobic and noninductive hydrophilic surfaces. When conidia were placed on hydrophobic coverslips, almost all ΔMorbp9 conidia germinated after 4 h, in a manner similar to those of wild type and Morbp9c conidia, suggesting that MoRBP9 is not required for conidial germination on hydrophobic surfaces. Surprisingly, these germinated conidia of ΔMorbp9 failed to develop appressoria at 8 h, or even at 24 h (Fig. 4). However, approximately 93.3% of conidia from the wild type developed appressoria, and a similar result was observed for Morbp9c, suggesting that MoRBP9 is required for appressorium development in response to hydrophobicity. The cAMP-dependent PKA signaling pathway has been shown to play a significant role in appressorium formation in M. oryzae (Li et al., 2017). We therefore applied exogenous cAMP to investigate whether the altered appressorium formation of ΔMorbp9 could be restored. Notably, approximately 52% and 16% of conidia from ΔMorbp9 were able to develop abnormally shaped appressoria after the addition of exogenous cAMP (2.5 mM) on hydrophobic and hydrophilic surfaces, respectively. These results showed that cAMP was capable of partially restoring appressorium formation in ΔMorbp9, indicating that MoRBP9 may be involved in the cAMP/PKA pathway.
M. oryzae also penetrates host plant cells via appressorium-like structures (ALS), which differentiate from the hyphal tip. We therefore formed ALS on hydrophobic coverslips to investigate whether MoRBP9 is involved in ALS development. The wild type formed mature ALS after 48 h, whereas ΔMorbp9 developed abnormal ALS (Fig. 5). Additionally, approximately 83.7% (type 2) of the ALS formed by ΔMorbp9 were irregular and unmelanized, and the remaining 16.3% (type 1) were smaller in size and less pigmented, suggesting that MoRBP9 regulates ALS morphology. Together, these results suggest that MoRBP9 is required for appressorium formation from conidial germ tubes and is involved in the morphology of ALS from hyphae.
Filamentous fungal hydrophobins confer hydrophobicity to the surface of conidia and mycelia. In M. oryzae, hydrophobins are involved in cell morphology and pathogenicity (Kim et al., 2005; Talbot et al., 1993; Wösten and Willey, 2000; Wösten et al., 1994). To investigate the functional roles of MoRBP9 in the hydrophobicity of M. oryzae, 5 μl drops of water or of 0.001% SDS and 0.025% SDS were placed on aerial hypha-bearing conidia. The results showed that water and SDS drops remained beaded on the aerial structures of the wild type (Fig. 6A). However, the water and SDS drops easily soaked ΔMorbp9, exhibiting a phenotype of easy wettability, whereas this defect was restored in Morbp9c. To further determine whether the hydrophobicity defect was related to altered expression of hydrophobin genes, we evaluated the expression of two well-characterized genes, MPG1 and MHP1, which regulate hydrophobicity in M. oryzae (Kim et al., 2005; Talbot et al., 1993). Compared with the wild type, MHP1 expression was not significantly affected in ΔMorbp9, whereas MPG1 expression was approximately 0.1-fold in ΔMorbp9, showing dramatically reduced expression (Fig. 6B). Overall, these results suggest that MoRBP9 plays an important role in the surface hydrophobicity of M. oryzae.
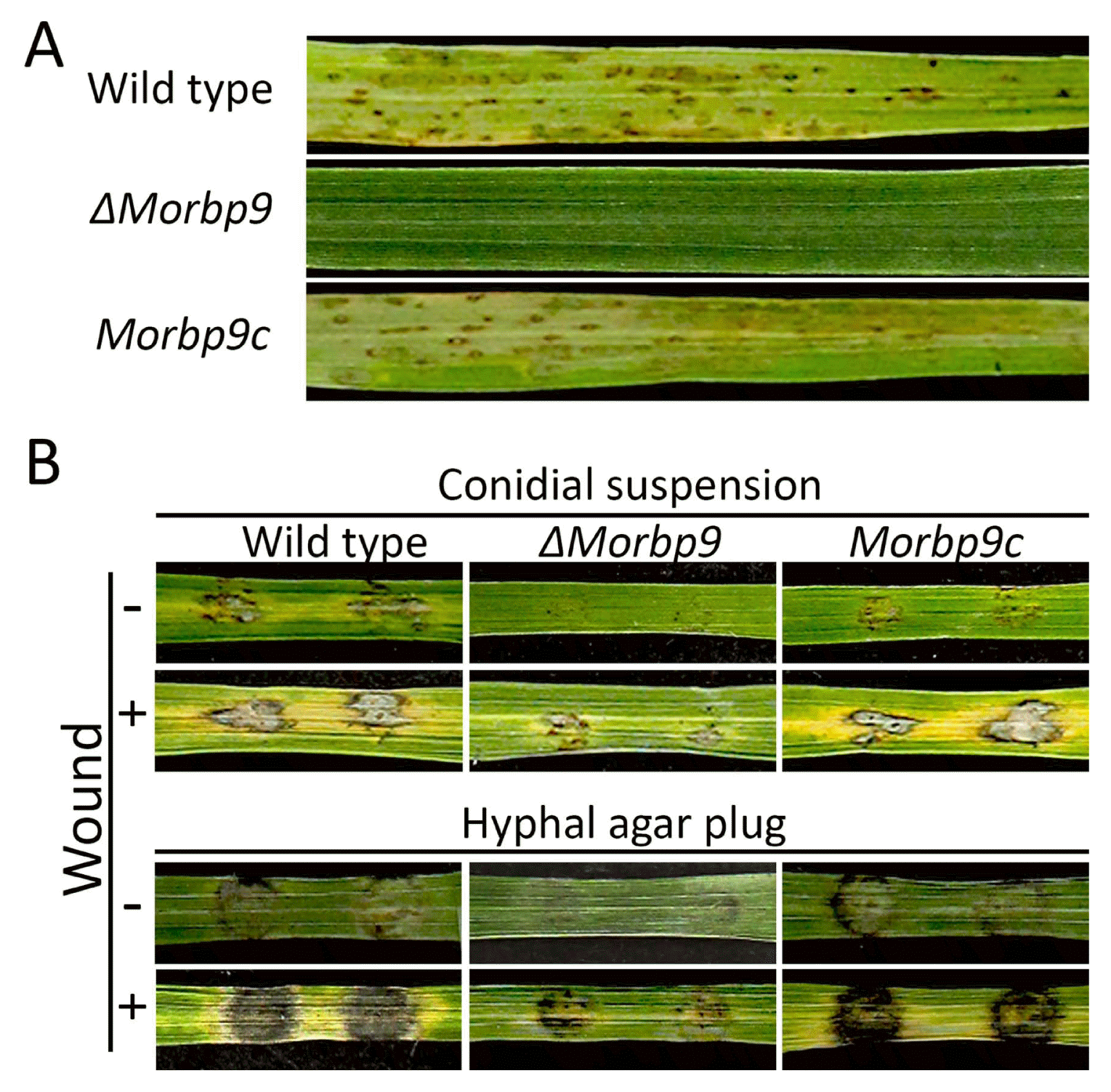
To assess the relationship between MoRBP9 and the pathogenicity of M. oryzae, 3-week-old susceptible rice seedlings were sprayed with a conidial suspension (105 conidia/ml). Wild type and Morbp9c caused numerous typical lesions, whereas ΔMorbp9 was nonpathogenic (Fig. 7A). To further evaluate the role of MoRBP9 in penetration of the rice epidermis, conidial drops were inoculated onto wounded and unwounded rice leaves. Consistent with the conidial spray assay, application of wild type and Morbp9c led to typical lesions on unwounded rice leaves, but ΔMorbp9 failed to cause any obvious lesions (Fig. 7B). However, ΔMorbp9 resulted in weaker lesions on wounded rice leaves compared with wild type and Morbp9c. Inoculation of hyphal plugs from ΔMorbp9 also caused weaker lesions on both wounded and unwounded rice leaves. Together, these results imply that MoRBP9 is involved in penetration and virulence.
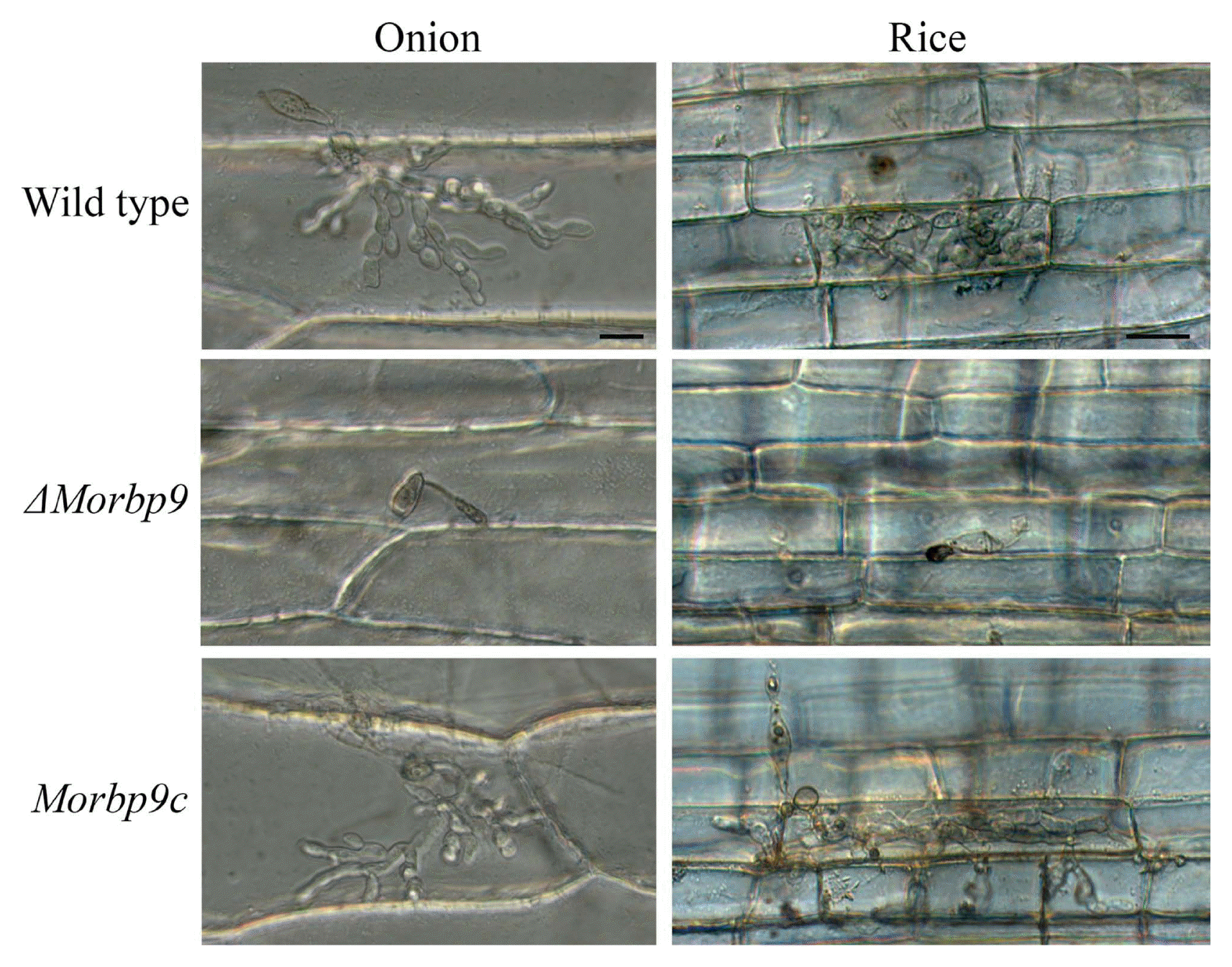
To further assess the role of MoRBP9 in pathogenic development, a conidial penetration assay was performed in onion epidermis and rice sheaths. Both wild type and Morbp9c formed invasive hyphae in onion and rice cells (Fig. 8). Unlike complete abolishment of appressorium formation on the hydrophobic surface, approximately 75% of ΔMorbp9 conidia produced abnormally shaped and less pigmented appressoria on the onion epidermis, which were unable to penetrate. On rice sheaths, approximately 80% of ΔMorbp9 conidia generated melanized appressoria, but they exhibited abnormal shapes and failed to penetrate rice sheath cells. Together, these results indicate that MoRBP9 regulates appressorium-mediated penetration and disease severity.
The microtubule-organization center, considered an important structure in which microtubule nucleation typically occurs, plays an important role in eukaryotic cell development. RanBPM, a putative scaffolding protein usually located in the microtubule-organization center, regulates many other proteins in multiple cellular processes (Salemi et al., 2017). The importance of RanBPM has been highlighted in previous studies (Salemi et al., 2017). Knockdown of RanBPM alters proper distribution of actin, which is important for forming a polarized network to allow movement of proteins and organelles (Etienne-Manneville, 2004; Kobayashi et al., 2007). Overexpression of RanBPM induces polarization of the mitochondrial membrane, resulting in activation of apoptosis by decreasing the protein level of B-cell lymphoma 2 (Bcl-2) and increasing bcl-2-like protein 4 (Bax) oligomerization (Liu et al., 2013). In co-immunoprecipitation and co-localization assays in live cells, RanBPM interacted with CDK11p46, which induces apoptosis (Mikolajczyk et al., 2003). RanBPM may be involved in transcriptional regulation via interaction with transcription factors such as homeobox A5, forkhead box, and high mobility group proteins (Atabakhsh et al., 2012). Additionally, transcriptional regulators such as steroid receptors, androgen receptors, glucocorticoid receptors, and TATA box binding protein-associated factor 4 have been reported to interact with RanBPM (Brunkhorst et al., 2005; Rao et al., 2002; Salemi et al., 2017). Although homologs of RanBPM were found to be evolutionarily conserved among eukaryotes, the functions of fungal RanBPM still remain largely unknown (Salemi et al., 2017). In the present study, we functionally characterized the RanBPM protein encoded by the MoRBP9 gene in M. oryzae and identified the external phenotypes related to pathogenesis. Phenotypical analyses revealed that loss of MoRBP9 resulted in developmental defects, including conidiation, appressorium formation, mycelial hydrophobicity, and virulence.
MoRBP9 was shown to regulate asexual reproduction, and deletion of MoRBP9 resulted in dramatic reductions in conidiation and conidiophore formation. We therefore concluded that MoRBP9 acts as a key regulator in the early stages of conidiation. In a previous study, 1,160 genes were found to be differently expressed during conidiation of M. oryzae in microarray analyses (Kim and Lee, 2012). Among the key regulatory genes, FLB3 and FLB4 were shown to control normal levels of mycelial formation and spore production, respectively, whereas COS1 regulated conidiophore formation (Matheis et al., 2017; Zhou et al., 2009). qRT-PCR analysis showed that these three genes were largely downregulated in ΔMorbp9 (Fig. 3). In particular, expression of COS1 in ΔMorbp9 was 0.25-fold of that in the wild type, indicating that MoRBP9 probably regulates the early stages of conidiation by acting as an upstream regulator of the expression of COS1, FLB3 and FLB4. However, it is still difficult to investigate the detailed mechanisms between MoRBP9 and these genes during the early stages of conidiation, because the specific functions of these genes in conidiophore formation are unknown. Preparing for conidiation, M. oryzae has been suggested to form a foot cell, which connects vegetative mycelium and aerial hyphae and provides nutrients and energy to aerial hyphae (He et al., 2013). The tips of aerial hyphae are remodeled to generate conidiophore by photoinduction (Bayry et al., 2012; Kim et al., 2009). Our qRT-PCR analysis showed that the expression level of MoRBP9 was similar between the conidiation and mycelium stages (Fig. 2). We speculated that the MoRBP9 protein may be translocated to aerial hyphae from vegetative mycelium and subsequently activated by some molecular signal related to conidiophore formation.
MoRBP9 is necessary for appressorium formation. Targeted deletion of MoRBP9 led to complete loss of appressorium formation and abnormally shaped ALS formation in response to hydrophobicity (Figs. 4 and 5). A previous study suggested that intracellular levels of RanBPM should be tightly manipulated, because different expression levels of RanBPM-encoded genes altered activation of the Ras/Erk pathway (Denti et al., 2004). In M. oryzae, the Ras/Erk pathway modulates appressorium development and subsequent infection (Jiang et al., 2018; Li et al., 2012). Considering that RanBPM contains only protein-protein interaction domains and functions as a scaffolding protein, it is reasonable to hypothesize that MoRBP9 acts as a key regulator of appressorium formation by modulating its partner proteins in signaling pathways such as the Ras/Erk pathway in M. oryzae, whereas ALS formation has been suggested to be involved in different mechanisms of appressoria (Kong et al., 2013). We suggest that MoRBP9 is involved in the morphology of ALS in M. oryzae. It is noteworthy that RanBPM indirectly maintains appropriate accumulation of intracellular cAMP levels in human kidney cells (Rex et al., 2010). We found that exogenous cAMP partly restored appressorium formation but failed to recover appressorium morphology, suggesting that MoRBP9 may affect other proteins involved in the cAMP-dependent PKA pathway. However, isolating specific partner proteins of RanBPM in this pathway is still challenging because of the large number of, and complex interactions among, these proteins. ΔMorbp9 still produced appressoria on the epidermis of rice and onion but failed to penetrate host cells, revealing that MoRBP9 is critical for appressorium-dependent penetration in M. oryzae (Fig. 8).
MoRBP9 is associated with the hydrophobicity of the aerial structures of M. oryzae. ΔMorbp9 was more hydrophilic compared with the wild type, revealed by placing drops of water or SDS solution on the aerial structures of M. oryzae (Fig. 6A). Like many other filamentous fungi, M. oryzae produces hydrophobins, which commonly coat the aerial mycelium and conidium (Han et al., 2015; Kim et al., 2005; Stringer et al., 1991; Talbot et al., 1993). In M. oryzae, mutation of MPG1 and MHP1, which encode class I and class II hydrophobins, respectively, resulted in defects in fungal development and pathogenicity (Kim et al., 2005; Talbot et al., 1993). However, these two mutants also exhibited distinct phenotypes such as conidial longevity and solvent-dependent hydrophilicity, suggesting different functions in MPG1 and MHP1 during development. It was shown that the expression levels of MPG1 were controlled by different regulators during development and under specific nutritional states (Soanes et al., 2002). Notably, in our study, expression of MPG1 but not MHP1 was largely reduced in the mycelia of ΔMorbp9 compared with the wild type (Fig. 6B). Our results therefore suggest that MoRBP9 is involved in the regulation of MPG1 expression to confer surface hydrophobicity in M. oryzae.
Acknowledgements
This study was supported by Basic Science Research Program through the National Research Foundation of Korea grant (NRF-2013R1A1A1008444) funded by the Ministry of Education, Science and Technology, Republic of Korea.
Notes
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Phylogenetic relationship, domain representation, and conserved amino acid sequence alignments of MoRBP9 and homologues from different organisms. (A) Phylogenetic analysis and domain representation of MoRBP9 and its homologues. A phylogenetic tree was generated by the neighbor-joining method with 1,000 bootstraps. The domain structures were predicted using InterProScan. SPRY, LIS1, CTLH and CRA indicates SPRY domain, LIS1 motif, CTLH motif and CRA domain, respectively. (B) Conserved amino acid sequence alignment of the domains. The identity of each protein from the BLAST search for MoRBP9 is followed by its name. Gray colored boxes indicate the domain and motif. Black boxes indicate that five amino acids were shared in the CTLH motif and CRA domain between Arabidopsis thaliana and Oryza sativa.
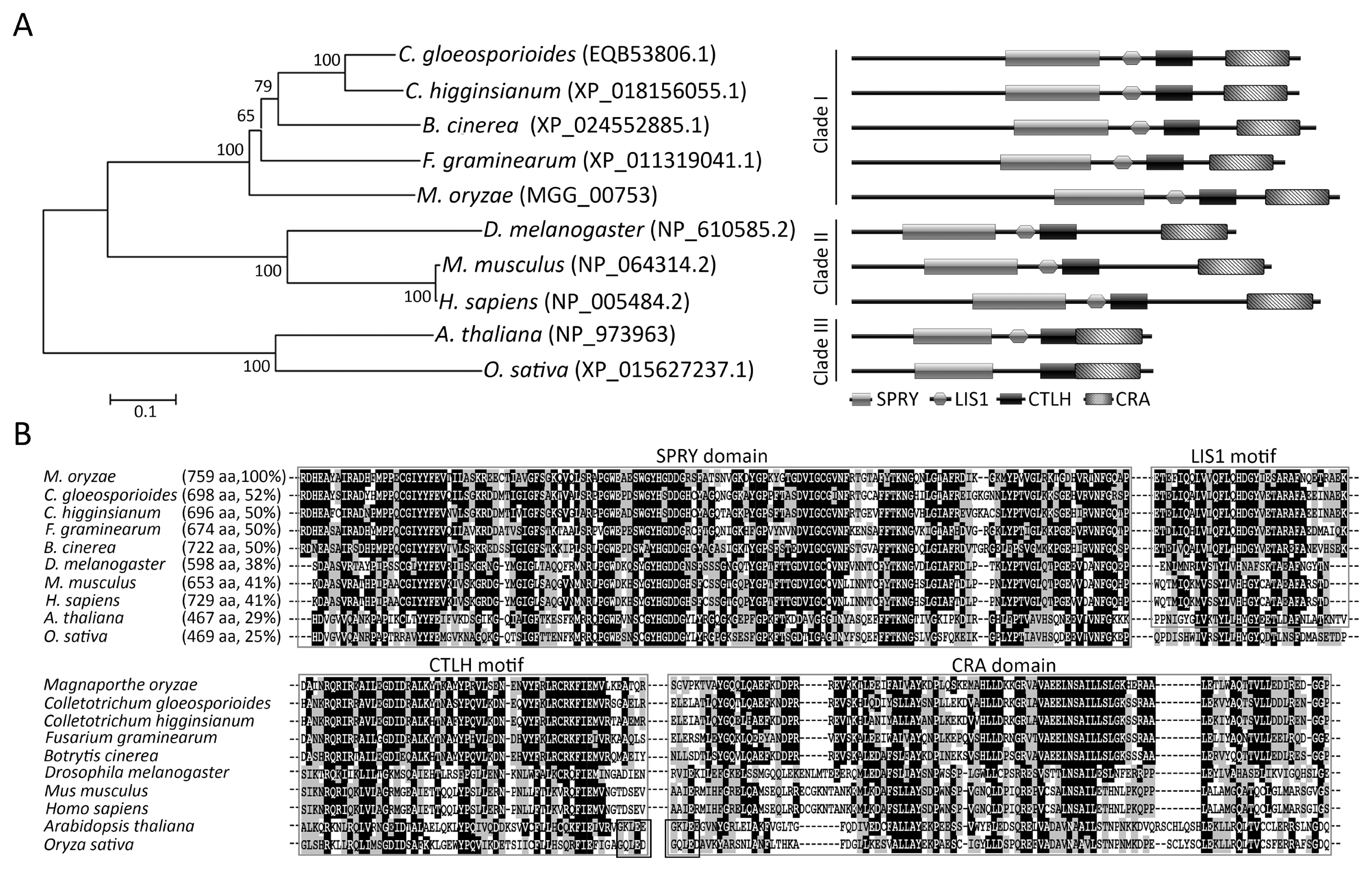
Fig. 2
Expression profiles and targeted deletion of MoRBP9. (A) The expression profile of MoRBP9 during different fungal developmental stages. Expression of MoRBP9 was measured by quantitative reverse transcription polymerase chain reaction. Total RNA was extracted from different fungal tissues containing mycelium (MY), conidium (CO), germinated conidium (GC), appressorium (AP), and infectious hyphae in rice leaves (IP). (B) Targeted deletion of MoRBP9. The deletion strategy was performed using the HPH cassette to replace MoRBP9 via the homologous recombination method. (C) Verification of MoRBP9 deletion by Southern blot analysis. Genomic DNA in the indicated strains was digested with HindIII and hybridized to a specific probe (approximately 500 bp in length). (D) Expression of MoRBP9 in the targeted deletion mutant. Expression of MoRBP9 was verified by reverse transcription polymerase chain reaction. Total RNA was extracted from the mycelia of wild-type, ΔMorbp9, and Morbp9c.
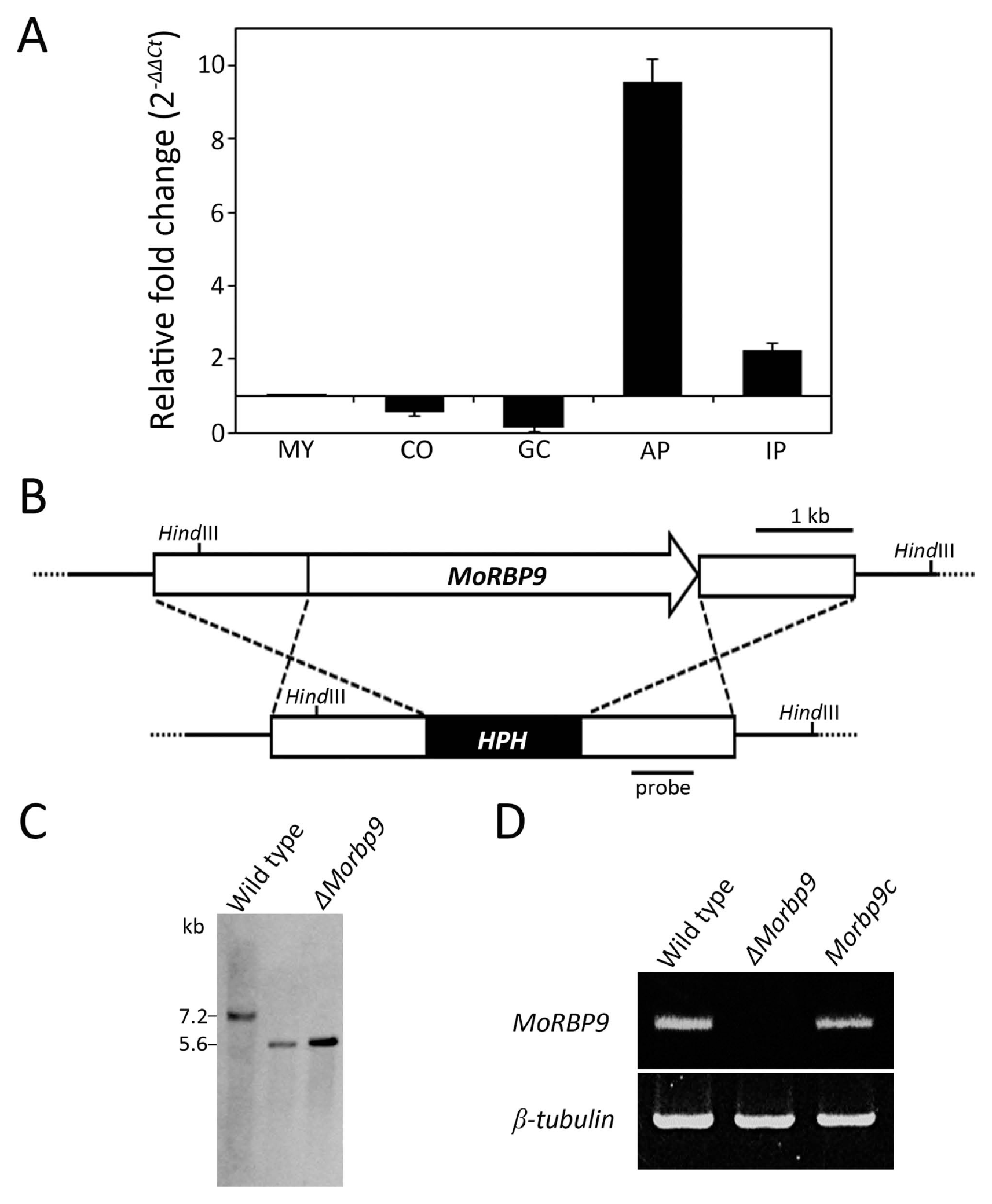
Fig. 3
Conidiation and conidiation-related gene expression. (A) Microscopic observation of conidiation. Microscopic observation was performed on strains grown on oatmeal agar (OMA) for 7 days. Scale bar = 20 μm. (B) Quantitative measurement of conidia. Conidia were collected from 7-day-old OMA in 5 ml distilled water. (C) Microscopic observations of conidiophore. Conidia and mycelia were stained with lactophenol blue solution and showed a blue color, while conidiophores showed no staining. Scale bar = 10 μm. (D) Expression of conidiation-related genes. Gene expression was measured by quantitative reverse transcription polymerase chain reaction. Total RNA was extracted from mycelia-bearing conidia.

Fig. 4
Appressorium formation on artificial surfaces. (A) Photographs of appressorium formation. Appressoria were formed on hydrophobic coverslips and hydrophilic slide glasses with or without the addition of exogenous cAMP (2.5 mM) after 8 h of incubation. Scale bar = 20 μm. (B) Statistical analysis of appressorium formation after 24 h.

Fig. 5
Appressorium-like structure (ALS) formation on hydrophobic surfaces. The ALS formation was seen on hydrophobic coverslips after incubation for 24 h. ΔMorbp9 was found to produce type 1 ALS (16.3%) and type 2 ALS (83.7%). Scale bar = 20 μm.
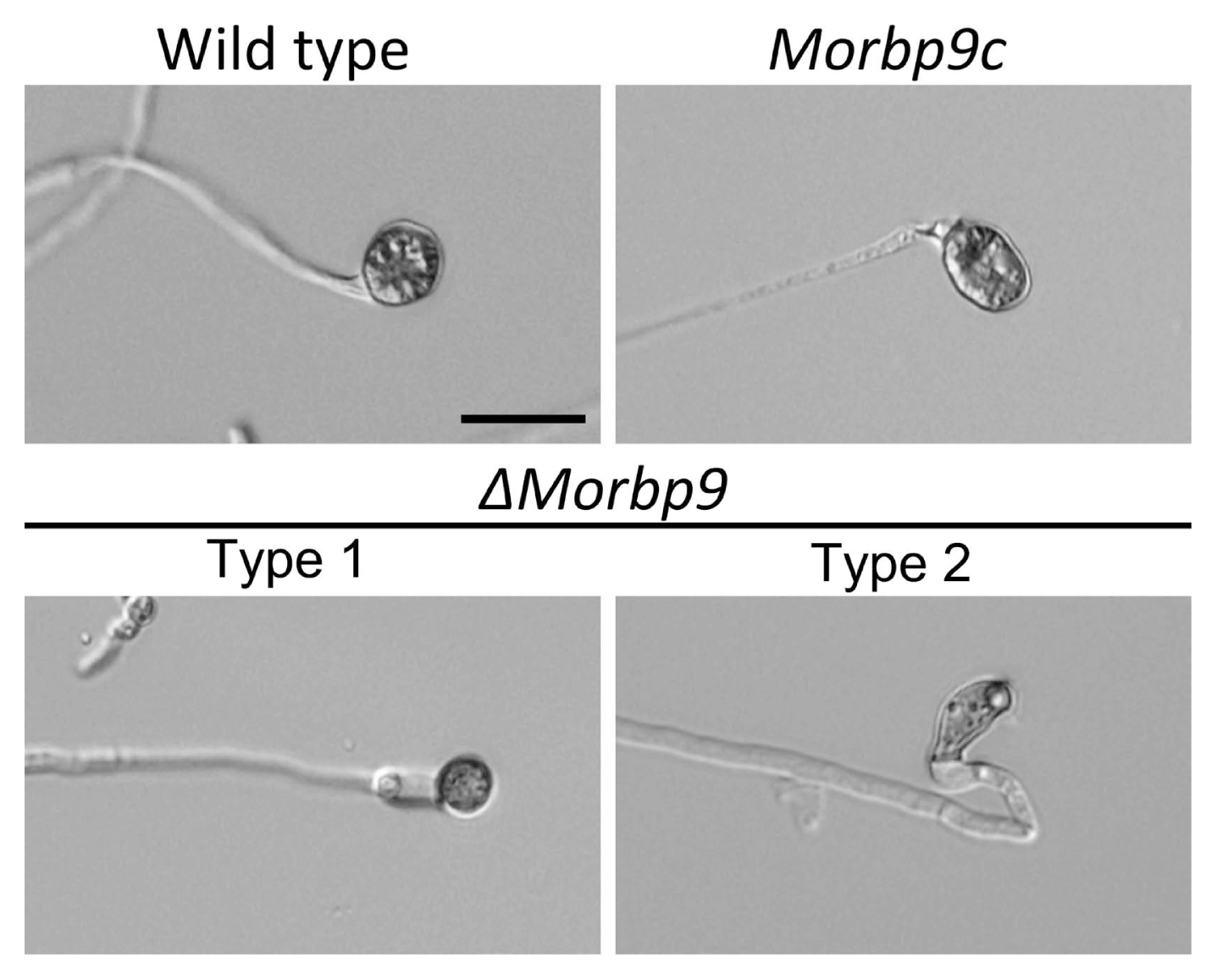
Fig. 6
Assessment of surface hydrophobicity. (A) The surface wettability of mycelial mats. Drops (10 μl) of distilled water, or 0.001% and 0.025% sodium dodecyl sulfate (SDS), were placed on aerial structures of the indicated strains cultured on oatmeal agar for 2 weeks. (B) The expression profiles of genes regulating hydrophobicity. Total RNA was extracted from mycelia cultured under the same conditions.
Fig. 7
Plant pathogenic assays. (A) Conidial spray assay. The conidial suspension was sprayed on rice leaves, which were incubated for 7 days at 25°C. (B) Disease development on wounded or unwounded rice leaves. Conidial drops and mycelial plugs were placed on wounded or unwounded rice leaves and incubated for 6 days.
References
Atabakhsh, E., Wang, J.H., Wang, X., Carter, D.E. and Schild-Poulter, C. 2012. RanBPM expression regulates transcriptional pathways involved in development and tumorigenesis. Am J Cancer Res. 2:549-565.


Bayry, J., Aimanianda, V., Guijarro, J.I., Sunde, M. and Latgé, J.-P. 2012. Hydrophobins-unique fungal proteins. PLoS Pathog. 8:e1002700



Brunkhorst, A., Karlén, M., Shi, J., Mikolajczyk, M., Nelson, M.A., Metsis, M. and Hermanson, O. 2005. A specific role for the TFIID subunit TAF4 and RanBPM in neural progenitor differentiation. Mol Cell Neurosci. 29:250-258.


Chi, M.-H., Park, S.-Y. and Lee, Y.-H. 2009. A quick and safe method for fungal DNA extraction. Plant Pathol J. 25:108-111.

Das, S., Haq, S. and Ramakrishna, S. 2018. Scaffolding protein RanBPM and its interactions in diverse signaling pathways in health and disease. Discov Med. 25:177-194.

Denti, S., Sirri, A., Cheli, A., Rogge, L., Innamorati, G., Putignano, S., Fabbri, M., Pardi, R. and Bianchi, E. 2004. RanBPM is a phosphoprotein that associates with the plasma membrane and interacts with the integrin LFA-1. J Biol Chem. 279:13027-13034.


Etienne-Manneville, S. 2004. Actin and microtubules in cell motility: which one is in control? Traffic. 5:470-477.


Fu, T., Kim, J.-O., Han, J.-H., Gumilang, A., Lee, Y.-H. and Kim, K.S. 2018. A small GTPase RHO2 plays an important role in pre-infection development in the rice blast pathogen Magnaporthe oryzae. Plant Pathol J. 34:470-479.



Han, J.-H., Lee, H.-M., Shin, J.-H., Lee, Y.-H. and Kim, K.S. 2015. Role of the MoYAK 1 protein kinase gene in Magnaporthe oryzae development and pathogenicity. Environ Microbiol. 17:4672-4689.


Han, J.-H., Shin, J.-H., Lee, Y.-H. and Kim, K.S. 2018. Distinct roles of the YPEL gene family in development and pathogenicity in the ascomycete fungus Magnaporthe oryzae. Sci Rep. 8:14461




He, Y., Deng, Y.Z. and Naqvi, N.I. 2013. Atg24-assisted mitophagy in the foot cells is necessary for proper asexual differentiation in Magnaporthe oryzae. Autophagy. 9:1818-1827.


Jiang, C., Zhang, X., Liu, H. and Xu, J.-R. 2018. Mitogen-activated protein kinase signaling in plant pathogenic fungi. PLoS Pathog. 14:e1006875



Kim, K.S. and Lee, Y.-H. 2012. Gene expression profiling during conidiation in the rice blast pathogen Magnaporthe oryzae. PLoS ONE. 7:e43202



Kim, S., Ahn, I.-P., Rho, H.-S. and Lee, Y.-H. 2005. MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol Microbiol. 57:1224-1237.


Kim, S., Park, S.-Y., Kim, K.S., Rho, H.-S., Chi, M.-H., Choi, J., Park, J., Kong, S., Park, J., Goh, J. and Lee, Y.-H. 2009. Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet. 5:e1000757



Kobayashi, N., Yang, J., Ueda, A., Suzuki, T., Tomaru, K., Takeno, M., Okuda, K. and Ishigatsubo, Y. 2007. RanBPM, Muskelin, p48EMLP, p44CTLH, and the armadillo-repeat proteins ARMC8α and ARMC8β are components of the CTLH complex. Gene. 396:236-247.


Kong, L.-A., Li, G.-T., Liu, Y., Liu, M.-G., Zhang, S.-J., Yang, J., Zhou, X.-Y., Peng, Y.-L. and Xu, J.-R. 2013. Differences between appressoria formed by germ tubes and appressorium-like structures developed by hyphal tips in Magnaporthe oryzae. Fungal Genet Biol. 56:33-41.


Leung, H., Lehtinen, U., Karjalainen, R., Skinner, D., Tooley, P., Leong, S. and Ellingboe, A. 1990. Transformation of the rice blast fungus Magnaporthe grisea to hygromycin B resistance. Curr Genet. 17:409-411.


Li, G., Zhou, X. and Xu, J.-R. 2012. Genetic control of infection-related development in Magnaporthe oryzae. Curr Opin Microbiol. 15:678-684.


Li, Y., Que, Y., Liu, Y., Yue, X., Meng, X., Zhang, Z. and Wang, Z. 2015. The putative Gγ subunit gene MGG1 is required for conidiation, appressorium formation, mating and pathogenicity in Magnaporthe oryzae. Curr Genet. 61:641-651.


Li, Y., Zhang, X., Hu, S., Liu, H. and Xu, J.-R. 2017. PKA activity is essential for relieving the suppression of hyphal growth and appressorium formation by MoSfl1 in Magnaporthe oryzae. PLoS Genet. 13:e1006954



Liu, T., Roh, S.E., Woo, J.A., Ryu, H. and Kang, D.E. 2013. Co-operative role of RanBP9 and P73 in mitochondria-mediated apoptosis. Cell Death Dis. 4:e476



Liu, W., Xie, S., Zhao, X., Chen, X., Zheng, W., Lu, G., Xu, J.-R. and Wang, Z. 2010. A homeobox Gene is essential for conidiogenesis of the rice blast fungus Magnaporthe oryzae. Mol Plant-Microbe Interact. 23:366-375.


Livak, K.J. and Schmittgen, T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 25:402-408.


Lu, J.-P., Feng, X.-X., Liu, X.-H., Lu, Q., Wang, H.-K. and Lin, F.-C. 2007. Mnh6, a nonhistone protein, is required for fungal development and pathogenicity of Magnaporthe grisea. Fungal Genet Biol. 44:819-829.


Matheis, S., Yemelin, A., Scheps, D., Andresen, K., Jacob, S., Thines, E. and Foster, A.J. 2017. Functions of the Magnaporthe oryzae Flb3p and Flb4p transcription factors in the regulation of conidiation. Microbiol Res. 196:106-117.


Mikolajczyk, M., Shi, J., Vaillancourt, R.R., Sachs, N.A. and Nelson, M. 2003. The cyclin-dependent kinase 11p46 isoform interacts with RanBPM. Biochem Biophys Res Commun. 310:14-18.


Nakamura, M., Masuda, H., Horii, J., Kuma, K.-I., Yokoyama, N., Ohba, T., Nishitani, H., Miyata, T., Tanaka, M. and Nishimoto, T. 1998. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to γ-tubulin. J Cell Biol. 143:1041-1052.



Nishitani, H., Hirose, E., Uchimura, Y., Nakamura, M., Umeda, M., Nishii, K., Mori, N. and Nishimoto, T. 2001. Full-sized RanBPM cDNA encodes a protein possessing a long stretch of proline and glutamine within the N-terminal region, comprising a large protein complex. Gene. 272:25-33.


Odenbach, D., Breth, B., Thines, E., Weber, R.W.S., Anke, H. and Foster, A.J. 2007. The transcription factor Con7p is a central regulator of infection-related morphogenesis in the rice blast fungus Magnaporthe grisea. Mol Microbiol. 64:293-307.


Park, J., Kong, S., Kim, S., Kang, S. and Lee, Y.-H. 2014. Roles of forkhead-box transcription factors in controlling development, pathogenicity, and stress response in Magnaporthe oryzae. Plant Pathol J. 30:136-150.



Perfetto, L., Gherardini, P.F., Davey, N.E., Diella, F., Helmer-Citterich, M. and Cesareni, G. 2013. Exploring the diversity of SPRY/B30.2-mediated interactions. Trends Biochem Sci. 38:38-46.


Qi, Z., Wang, Q., Dou, X., Wang, W., Zhao, Q., Lv, R., Zhang, H., Zheng, X., Wang, P. and Zhang, Z. 2012. MoSwi6, an APSES family transcription factor, interacts with MoMps1 and is required for hyphal and conidial morphogenesis, appressorial function and pathogenicity of Magnaporthe oryzae. Mol Plant Pathol. 13:677-689.



Rao, M.A., Cheng, H., Quayle, A.N., Nishitani, H., Nelson, C.C. and Rennie, P.S. 2002. RanBPM, a nuclear protein that interacts with and regulates transcriptional activity of androgen receptor and glucocorticoid receptor. J Biol Chem. 277:48020-48027.


Rex, E.B., Rankin, M.L., Yang, Y., Lu, Q., Gerfen, C.R., Jose, P.A. and Sibley, D.R. 2010. Identification of RanBP 9/10 as interacting partners for protein kinase C (PKC) γ/δ and the D1 dopamine receptor: regulation of PKC-mediated receptor phosphorylation. Mol Pharmacol. 78:69-80.



Sakulkoo, W., Osés-Ruiz, M., Garcia, E.O., Soanes, D.M., Littlejohn, G.R., Hacker, C., Correia, A., Valent, B. and Talbot, N.J. 2018. A single fungal MAP kinase controls plant cell-tocell invasion by the rice blast fungus. Science. 359:1399-1403.


Salemi, L.M., Maitland, M.E.R., McTavish, C.J. and Schild-Poulter, C. 2017. Cell signalling pathway regulation by RanBPM: molecular insights and disease implications. Open Biol. 7:170081



Shi, Z. and Leung, H. 1995. Genetic analysis of sporulation in Magnaporthe grisea by chemical and insertional mutagenesis. Mol Plant-Microbe Interact. 8:949-959.

Soanes, D.M., Kershaw, M.J., Cooley, R.N. and Talbot, N.J. 2002. Regulation of the MPG1 hydrophobin gene in the rice blast fungus Magnaporthe grisea. Mol Plant-Microbe Interact. 15:1253-1267.


Stringer, M.A., Dean, R.A., Sewall, T.C. and Timberlake, W.E. 1991. Rodletless, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 5:1161-1171.


Talbot, N.J. 2003. On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu Rev Microbiol. 57:177-202.


Talbot, N.J., Ebbole, D.J. and Hamer, J.E. 1993. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 5:1575-1590.



Thines, E., Weber, R.W.S. and Talbot, N.J. 2000. MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell. 12:1703-1718.



Tomaštíková, E., Cenklová, V., Kohoutová, L., Petrovská, B., Váchová, L., Halada, P., Kočárová, G. and Binarová, P. 2012. Interactions of an Arabidopsis RanBPM homologue with LisH-CTLH domain proteins revealed high conservation of CTLH complexes in eukaryotes. BMC Plant Biol. 12:83



Wang, D., Li, Z., Schoen, S.R., Messing, E.M. and Wu, G. 2004. A novel MET-interacting protein shares high sequence similarity with RanBPM, but fails to stimulate MET-induced Ras/ Erk signaling. Biochem Biophys Res Commun. 313:320-326.


Wösten, H.A.B., Asgeirsdóttir, S.A., Krook, J.H., Drenth, J.H. and Wessels, J.G. 1994. The fungal hydrophobin Sc3p selfassembles at the surface of aerial hyphae as a protein membrane constituting the hydrophobic rodlet layer. Eur J Cell Biol. 63:122-129.

Wösten, H.A.B. and Willey, J.M. 2000. Surface-active proteins enable microbial aerial hyphae to grow into the air. Microbiology. 146:767-773.


Yang, J., Zhao, X., Sun, J., Kang, Z., Ding, S., Xu, J.-R. and Peng, Y.-L. 2010. A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae. Mol Plant-Microbe Interact. 23:112-123.


Yi, M., Park, J.-H., Ahn, J.-H. and Lee, Y.-H. 2008. MoSNF1 regulates sporulation and pathogenicity in the rice blast fungus Magnaporthe oryzae. Fungal Genet Biol. 45:1172-1181.





 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




