Actin Cytoskeleton and Golgi Involvement in Barley stripe mosaic virus Movement and Cell Wall Localization of Triple Gene Block Proteins
Article information
Abstract
Barley stripe mosaic virus (BSMV) induces massive actin filament thickening at the infection front of infected Nicotiana benthamiana leaves. To determine the mechanisms leading to actin remodeling, fluorescent protein fusions of the BSMV triple gene block (TGB) proteins were coexpressed in cells with the actin marker DsRed: Talin. TGB ectopic expression experiments revealed that TGB3 is a major elicitor of filament thickening, that TGB2 resulted in formation of intermediate DsRed:Talin filaments, and that TGB1 alone had no obvious effects on actin filament structure. Latrunculin B (LatB) treatments retarded BSMV cell-to-cell movement, disrupted actin filament organization, and dramatically decreased the proportion of paired TGB3 foci appearing at the cell wall (CW). BSMV infection of transgenic plants tagged with GFP-KDEL exhibited membrane proliferation and vesicle formation that were especially evident around the nucleus. Similar membrane proliferation occurred in plants expressing TGB2 and/or TGB3, and DsRed: Talin fluorescence in these plants colocalized with the ER vesicles. TGB3 also associated with the Golgi apparatus and overlapped with cortical vesicles appearing at the cell periphery. Brefeldin A treatments disrupted Golgi and also altered vesicles at the CW, but failed to interfere with TGB CW localization. Our results indicate that actin cytoskeleton interactions are important in BSMV cell-to-cell movement and for CW localization of TGB3.
Intracellular transport of plant viruses during infection operates via unique interactions between viral proteins and selected host cytoskeletal and membrane elements. Micro-tubules and/or actin filaments are required for host cyto-skeleton functions that are involved in virus replication and movement (Amari et al., 2011). Virus infections often result in modification of these systems and mechanisms affecting these processes have become special topics of enquiry over the past decade (Ueki and Citovsky, 2011). For example, the roles of cellular remodeling in formation of replication factories and intracellular movement of several viruses has been reviewed recently (Laliberte and Safacon, 2010; Schoelz et al., 2011). Since these reviews, the Potato virus X (PVX) triple gene block 1 (TGB1) movement protein has been shown to remodel actin and endomembranes to form X-bodies that function in replication, and to recruit the TGB2 and TGB3 proteins to the X-bodies (Tilsner et al., 2012). Also, the endoplasmic reticulum (ER) and Golgi apparatus have been shown to be extensively remodeled as a consequence of Turnip mosaic virus (TuMV) infection (Grangeon et al., 2012).
Modified actin filaments and the endomembrane system are involved intracellular and intercellular movement of several plant viruses (Hawes and Satiat-Jeuenemairte, 2005; Ju et al., 2005; Lucas, et al., 2009; Vogel et al., 2007; Wright et al., 2007). The plasma membrane and ER are continuous between cells, and form desmotubule conduits for inter-cellular movement of macromolecules between adjacent cells that are regulated by actin filaments, myosin-motor associations, and/or poorly understood membrane flow mechanisms (Epel, 2009; Niehl and Heinlein, 2011; Ritzenthaler, 2011; Schoelz et al., 2011). Virus studies over the past decade have shown that plasmodesmata (PD) connections forming cellular symplast boundaries are remarkably plastic and are involved in numerous complex interactions required for virus transit (Maule et al., 2011; Ueki and Citovsky, 2011). Considerable variation is now apparent in mechanisms of virus intercellular movement, and this is most evident in viruses in which nucleoprotein complexes move through enlarged PD, versus viruses in which remodeled tubular PD function in cell-to-cell transit of whole virions (Epel, 2009; Maule et al., 2011; Scholthof, 2005; Ueki and Citovsky, 2011).
All plant viruses encode movement proteins (MP) that function in cell-to-cell movement, but these proteins and the pathways involved in movement vary enormously in their complexity and host component interactions (Schoelz et al., 2011; Verchot, 2011). The most intensively studied MPs are Tobacco mosaic virus (TMV) 30K protein, TGB proteins encoded by hordeiviruses and potexviruses, the Closterovirus movement complex proteins, the Nepovirus 2B protein, and the P6 Cauliflower mosaic virus (CaMV) (Schoelz et al., 2011; Verchot, 2011). Each of these MP complexes interact in a variety of ways with endomembrane components, cytoskeleton networks and PD (Amari et al., 2011; Ritzenthaler, 2011; Schoelz et al., 2011; Verchot et al., 2010). Even closely related viruses sometimes vary in their mechanisms of movement; for example TMV localized movement is strongly affected by actin filament disruption, whereas the closely related Turnip vein clearing virus (TVCV) is insensitive to actin disruption and has been hypothesized to move primarily by membrane flow rather than by actin cytoskeleton interactions (Harries et al., 2009; Harries et al., 2010; Schoelz et al., 2011). Hence, it is important to carry out analyses with a wide range of diverse viruses and virus strains to obtain a more complete view of the varied mechanisms culminating in local and systemic movement.
The focus of our studies over the past few years has involved movement of Barley stripe mosaic virus (BSMV), the type member of the Hordeivirus genus, whose members encode TGB movement proteins and have been classified in the rod-shaped Virgaviridae family (Jackson et al., 2009: Verchot, 2011). Members of the Hordeiviruses have positive-sense, single-stranded RNA genomes consisting of three segments designated to α, β, and γ, and they differ in this respect from PVX, a monopartite member of the Flexiviridae, whose TGB movement functions have also been intensively investigated (See Verchot et al., 2010, and references therein). The Hordeivirus genus contains Poa semilatent virus (PSLV), Lychnis ring spot virus (LRSV) and Anthoxanthum latent blanching virus (ALBV), and amongst these, the properties of PSLV TGB proteins have been investigated intensively by the Morozov group (Jackson et al., 2009; Verchot et al., 2010). Hordeivirus RNAβ segments encode the coat protein (CP), which is translated directly from the genomic (g) RNA, and the TGB proteins, which are expressed from two subgenomic (sg) RNAs (Zhou and Jackson, 1996). The CP is dispensable for systemic movement of BSMV, but not PVX, and mutational analyses of BSMV indicate that the TGB1, TGB2 and TGB3 proteins are essential for cell-to-cell movement in both monocot and dicot hosts (Lawrence and Jackson, 2001a). Previously, we showed that TGB3 interacts physically with both TGB1 and TGB2 and that these interactions are required for virus cell-to-cell movement and for targeting of both proteins to the PD (Lim et al., 2008; Lim et al., 2009). In a more distantly related member of the Virgaviridae family, Potato mop-top virus (PMTV), interactions between TGB2 and TGB3 are also involved in TGB1 localization and movement (Cowan et al., 2002; Shemyakina et al., 2011; Zamyatnin et al., 2004). In addition, BSMV TGB1 has a variety of biochemical functions and forms nucleoprotein complexes with viral RNA in vivo that appear to function in cell-to-cell transit (Lim et al., 2008). Separate findings also reveal that interactions of PSLV and PMTV TGB2 and TGB3 function to increase the size-exclusion limits of PD, and questions have arisen as to whether that the TGB2 protein associates with TGB3 to move to adjacent cells through actin-ER network associations during transit to PD, or is transported by other mechanisms (Haupt et al., 2005, Shemyakina et al., 2011; Verchot, 2011; Verchot-Lubicz et al., 2010). Our collective results suggest that a variety of mechanisms regulate TGB protein cellular interactions amongst viruses in the Virgaviridae family; hence a more complete understanding of the nuances of TGB viruses during movement requires additional approaches.
We previously evaluated the subcellular localization of the BSMV GFP (green fluorescent protein):TGB1 protein by inoculating plants and protoplasts with reporter virus mutants that affect the TGB protein functions (Lawrence and Jackson, 2001), and recently have conducted more detailed analyses of the roles of the TGB proteins in PD targeting by using an Agrobacterium-mediated transient protein expression system in N. benthamiana (Lim et al., 2008; Lim et al., 2009). Among the more important findings is evidence that BSMV TGB2 and TGB3 form ribonucleo-protein (RNP) complexes with TGB1 that may function during during cell-to-cell transport (Lim et al., 2008; Lim et al., 2009). We have now extended these investigations to assess associations of the TGB proteins with the cytoskeleton, membrane vesicles, and the Golgi apparatus of the host. Agrobacterium transient expression of GFP:Talin or DsRed: Talin in N. benthamiana plants provided markers for actin filaments (Lim et al., 2009). A GFP:KDEL fusion protein directed to ER (Gomord et al., 1997), a TuA:GFP marker for tubulin (Ueda et al., 1999), and an ST-GFP Golgi marker (Boevink et al., 1998) were used to visualize specific cellular components. The results indicate that during BSMV infection, actin filaments are remodeled to form thick cables and that the TGB3 protein has a major role in filament remodeling. TGB3 also has substantial roles in eliciting formation of membrane vesicles associated with the cell wall (CW), and the Golgi, and the targeting of actin to these structures.
Materials and Methods
In vitro transcription reactions and plant inoculation
Infectious BSMV α, β, γ, and γ: γb-GFP gRNA transcripts were produced by T7 RNA polymerase transcription of linearized cDNA templates as previously described (Petty et al., 1989). The transcribed gRNAs were mixed in equal proportions and mechanically inoculated to N. benthamiana as described (Lim et al., 2008).
Agrobacterium infiltration and BSMV cDNA constructs
The binary vectors (pGD, pGDG, and pGDR) expressing wild type (wt) TGB1, TGB2, TGB2/3, and TGB3, several TGB mutant derivatives, and a talin actin marker previously were used in transient expression experiments (Lim et al., 2008; Lim et al., 2009). Agrobacterium infiltrations of N. benthamiana leaves were performed essentially as described for “silencing on the spot” (Johansen and Carrington, 2001) with minor modifications that we have implemented previously (Lim et al., 2008). For agroin-filtration, bacteria harboring the desired TGB combinations were mixed with the silencing suppressor construct pGD:Yb (Bragg et al., 2004), and after a 2 to 3 h incubation of the mixtures at room temperature, leaves were pressure infiltrated with a 1 ml syringe. For plasmolysis before microscopic analysis, 700 mM sucrose was infiltrated as described by Lim et al. (2009). Transgenic N. benthamiana lines expressing the GFP-KDEL ER marker and the TuA-GFP microtubule marker were provided by David Baulcombe (John Innes Centre, Norwich, United Kingdom), and used to test associations with TGB proteins (Lim et al., 2008). The rat sialyl transferase GFP (ST-GFP) marker provided by Valerian Dolja (Oregon State University) was cloned into Agrobacterium and used to visualize Golgi membranes (Boevink et al., 1998).
Treatment with microfilament destabilizing drugs and Golgi inhibitors
For treatment of N. benthamiana, a stock solution of 10 mM Latrunculin B (LatB; Sigma) was dissolved in dimethylsulfoxide (DMSO). Prior to infiltration, the stock was diluted to 5 μM, 20 μM, or 50 μM LatB in Agrobacterium incubation mixture and an equivalent DMSO dilution was infiltrated as a buffer control (Lim et al., 2008). Several regions of the same leaf were treated by infiltration of the LatB or DMSO dilutions 4 h before virus inoculation. A 10 mg/ml stock solution of Brefeldin A (BFA) in DMSO (Sigma-Aldrich, London, UK) was diluted to a concentration of 100 μg/mL and infiltrated as described (daSilva et al., 2004).
Fluorescence assays
Confocal laser scanning microscopy with a Zeiss LSM 510 Meta microscope (Thornwood, New York) was employed to visualize GFP fluorescence in epidermal cells of N. benthamiana leaves. In order to detect GFP fluorescence, an Argon 488 nm laser and 505–530 nm band pass emission filter was used before transmission to a photomultiplier tube detector. DsRed excited with a HeNeI 543 nm laser was imaged using the META detector set for 570–600 nm. Calcofluor Blue stained walls were visualized by capture of emission spectra between 375 and 450 nm excited by an Enterprise UV laser set at 358 and 364 nm range, and the resulting fluorescence was digitally manipulated to appear white (Lim et al., 2008).
Results
Actin filament structure is altered in BSMV-infected N. benthamiana epidermal cells
Several plant virus movement proteins have been reported to interact with cyto-skeleton elements during cell-to-cell movement (Ferralli et al., 2006; Grangeon et al., 2012; Schoelz et al., 2011; Tilsner et al., 2012), and the MPs of TMV and Cucumber mosaic virus have been reported to cleave actin at the PD (Su et al., 2010). To determine whether BSMV affects actin cyto-skeleton patterns, N. benthamiana leaves were inoculated with wild-type (wt) BSMV α and β gRNAs, and gRNAγ containing the γb:GFP fusion protein as a marker for virus movement (Fig. 1A; green background). At 5 days post-inoculation (dpi), DsRed:Talin was co-expressed in the same leaves by agroinfiltration to permit visualization of actin filaments and possible changes to the cytoskeleton architecture as BSMV moved through the infiltrated region (Fig. 1A; red background). The results revealed that DsRed:Talin was converted into a dense cable-like pattern at the BSMV infection front (Fig. 1B) that was quite distinct from the filamentous profile observed in uninvaded tissue (Fig. 1C). Because these observations suggest that BSMV infection caused drastic changes to the cytoskeletal profile, we carried out experiments with the TGB proteins to determine whether they might be involved in triggering the cytoskeleton changes and to determine whether the changes affect cytoplasmic localization of the TGB proteins.

Changes in actin filament structure in BSMV-infected N. benthamiana epidermal cells. (A) N. benthamiana leaf inoculated with a BSMV derivative consisting of wild-type α and β RNAs and RNAγ containing the γb:GFP fusion protein as a marker for virus infection (green). DsRed:Talin was co-expressed in the same leaf by agroinfiltration to permit visualization of actin filaments (red). Cells were examined by confocal microscopy at 3 to 10 days after inoculation, and actin filament thickness was examined at the infection fronts of cells. (B) Thick remodeled actin cables visualized by DsRed:Talin (C) DsRed:Talin expression showing the filamentous actin network in an uninvaded region of the leaf. Note: Substantially thicker actin filaments were present only in regions at or behind the infection front of regions expressing BSMV GFP. Photographs were taken at 7 dpi.
TGB3 induces thickened actin cables similar to those in BSMV-infected tissue
To evaluate the ability of individual TGB proteins to induce actin filament modifications, TGB1, TGB2, and TGB3 were agroinfiltrated into N. benthamiana leaves along with bacteria harboring DsRed:Talin or GFP: Talin (Lim et al., 2008; Lim et al., 2009). When expressed alone, the DsRed:Talin protein associated with thin actin filaments distributed throughout the cell (Fig. 2A). GFP: TGB1 fluorescence did not typically colocalize with DsRed:Talin, but instead appeared to form large globular vesicles, and Z stack analysis suggested that the protein was localized relatively randomly in the cytosol (Fig. 2E). Although TGB1 had no obvious effects on the fluorescence patterns of DsRed:Talin (Fig. 2E), in tissue infiltrated with TGB2, the associated actin filaments formed “intermediate bundles” that were more distinct than those formed in leaves expressing TGB1. GFP:TGB2 also formed vesicle-like spots that were closely associated with the DsRed:Talin filaments (Fig. 2B). In contrast, ectopic expression of both the DsRed:TGB3 fusion protein (Fig. 2C) and wtTGB3 (data not shown) elicited thick DsRed:Talin cables resembling those at the BSMV infection front. Infiltrated bacteria harboring GFP:TGB2/3, which produces the ~10:1 ratio of TGB2 to TGB3 found in infected cells (Zhou and Jackson, 1996), also lead to the formation of thick actin cables and large vesicles containing GFP:TGB1 (Fig. 2D).

TGB2 and TGB3 induction of actin filament bundling and roles in TGB1 PD targeting. The actin marker protein, DsRed:Talin, was coexpressed in N. benthamiana leaves along with TGB proteins by agroinfiltration. The following combinations are shown: (A) DsRed:Talin (B) GFP:TGB2 + DsRed:Talin (C) DsRed:TGB3 + GFP:Talin (D) GFP:TGB2/3 + DsRed:Talin (E) GFP:TGB1 + DsRed:Talin (F) GFP:TGB1, TGB2 + DsRed:Talin (G) GFP:TGB1, TGB3 + DsRed:Talin (H) GFP:TGB1, TGB2/3 + DsRed:Talin. In (E-H) the top panels represent single confocal images, and the bottom panels represent a Z-stack projection of over 32 optical slices through the cell generated using the Imaris software. Bars = 20 μm. Photographs were taken at 2 days post infiltration.
We previously showed that trafficking of TGB1 to the PD depends on expression of TGB3 (Lim et al., 2009). To evaluate TGB1 associations with actin filaments in the presence of other TGB proteins, GFP:TGB1 and DsRed: Talin were co-expressed with TGB2 and TGB3 or with TGB2/3. As noted above, when expressed in the absence of TGB2 or TGB3, GFP:TGB1 appeared in large vesicles dispersed throughout the cell that did not associate closely with the thin DsRed:Talin filaments (Fig. 2E). After co-expression with TGB2, GFP:TGB1 localized in larger vesicle-like structures with an apparently random distribution, but the actin network formed intermediate filaments that were more distinct than those observed when GFP: TGB1 was co-expressed with DsRed:Talin (Fig. 2F). As anticipated from Fig. 1, cells coexpressing GFP:TGB1 and TGB3, DsRed:Talin formed thickened cables typical of those at the front of BSMV invaded tissue, and these often overlapped with the GFP:TGB1 fluorescence (Fig. 2G). When GFP:TGB1 was paired with the TGB2/3 construct for coexpression of all three TGB proteins, TGB1 co-localized with the cables in vesicle-like spots that were smaller than those observed during coexpression with either TGB2 or TGB3 alone (Fig. 2H). In addition, the Z stack compilations suggested that GFP:TGB1 was more closely associated with the cell wall (CW) of cells coexpressing TGB3 or TGB2/3 than when expressed in the absence of TGB3. Together these results show that TGB3, and to a lesser extent TGB2, induces actin cytoskeleton network thickening, and suggest that the relatively low expression of TGB3 from the TGB2/3 construct is sufficient to mediate this process.
TGB2, TGB3 and TGB2/3 are associated with ER and actin filaments
To extend our earlier observations that BSMV protoplast infections result in membrane proliferation (Lawrence and Jackson, 2001b), we infected transgenic N. benthamiana expressing the ER marker, GFP-KDEL (Fig. 3). Fluorescence microscopy of uninfected leaf cells revealed an array of membranes of various compositions (Fig. 3A, top) that often had a reticulate appearance (Fig. 3A, bottom). Observation of BSMV infected cells at 7 dpi indicated a substantial increase in membrane density, particularly around the nuclei (Fig. 3B, top), and higher magnification revealed globular vesicles surrounding the nuclei (Fig. 3B, bottom). To extend these observations, and to determine whether transiently expressed TGB proteins localized in the vesicles, various TGB combinations were transiently expressed from pGD plasmids by agroinfiltration of the GFP-KDEL plants. Individual leaves from the transgenic plants were divided into eight regions and each region was infiltrated with Agrobacterium to mediate expression of DsRed:Talin, and one of the following combinations: a pGD vector control, TGB1, TGB2, TGB3, or TGB2/3 (Fig. 4A). The ER of leaf sections infiltrated with the pGD vector control formed a compact network that colocalized with thin, high density actin filaments (Fig. 4B), and in regions expressing the TGB1 protein, the ER/actin network was similar to that observed in the vector control regions (Fig. 4C). The TGB2 treated regions had a more obvious reticulate membrane pattern, but otherwise the membranes appeared similar to those observed in the control and TGB1 infiltrated regions (Fig. 4D). In contrast, the ER network was altered substantially in regions expressing the TGB3, and TGB2/3 proteins, and consisted of numerous small, vesicles associated with thickened actin cables (Fig. 4E and 4F). Expression of TGB3 induced a large number of vesicles, and the changes in the ER/actin network appeared to be substantially more pronounced in these sections than in regions expressing TGB2/3 (Fig. 4E). These results indicate that both TGB2 and TGB3 contribute to membrane proliferation during transient expression and that some elicited vesicles co-localize with remodeled actin filaments.
Vesicle appearance in BSMV infected GFP-KDEL transgenic N. benthamiana epidermal cells. (A) Uninfected GFP-KDEL N. benthamiana. (B) N. benthamiana leaf inoculated with BSMV α, β, and γ RNAs. Arrows indicate vesicle formation around nucleus as a marker for virus infection (green). Cells were examined by confocal microscopy at 7 days after inoculation. Bars = 50 μm. The magnification of the exploded bottom panel exactly repeats the boxed areas in the upper panels.
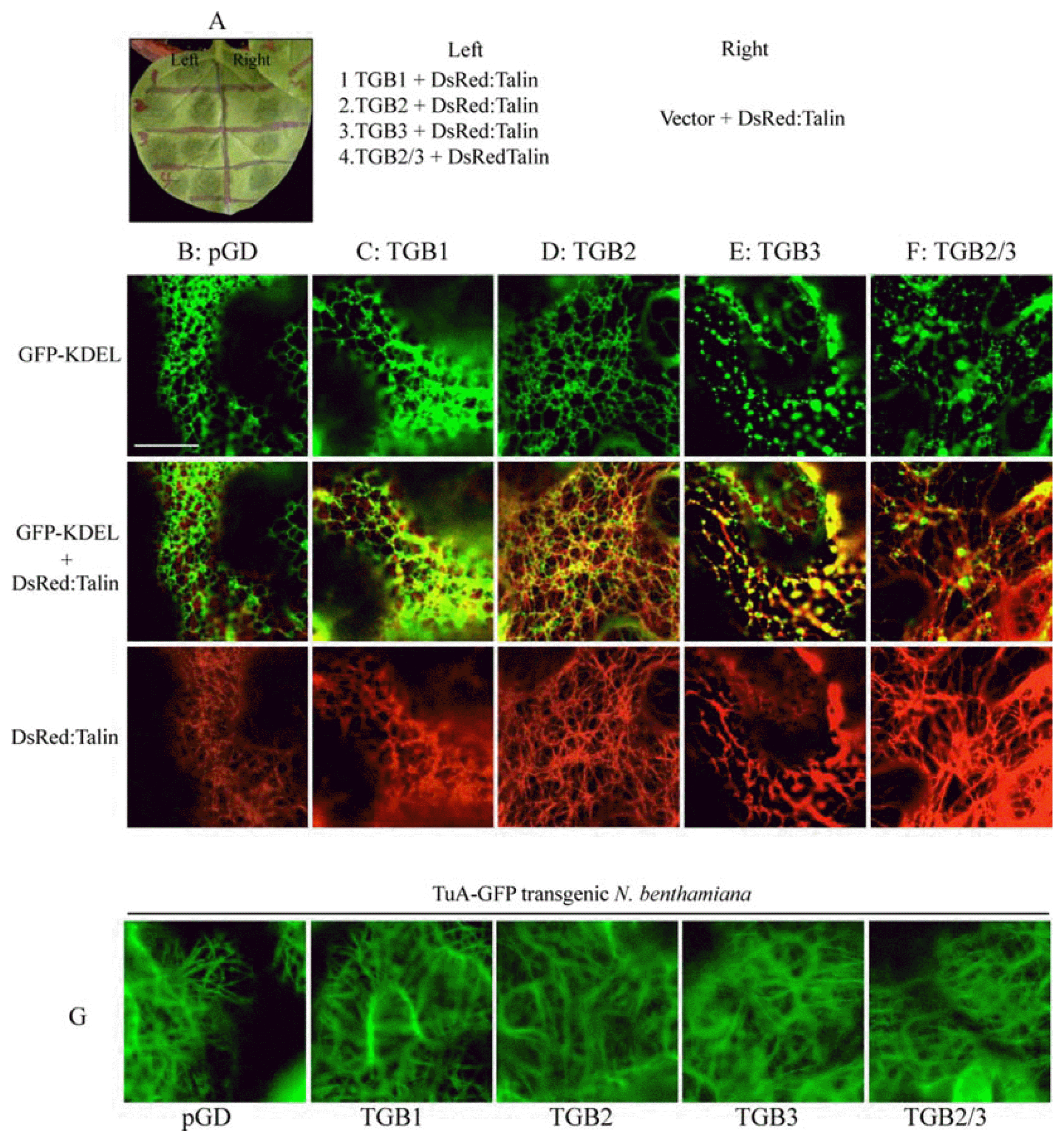
TGB2 and TGB3 effects on ER-derived vesicles. The actin marker protein, DsRed:Talin, was expressed with the TGB proteins by agroinfiltration into GFP-KDEL transgenic N. benthamiana (A) In order to compare the effects of the TGB proteins on ER and actin within same leaf, the leaf was divided into eight sections and infiltrated with the following combinations: (B) Agrobacterium pGD vector + DsRed:Talin (C) TGB1 + DsRed:Talin (D) TGB2 + DsRed:Talin, (E) TGB3 + DsRed:Talin, or (F) TGB2/3 + DsRed:Talin. (B–F) (First panel) green channel shows GFP-KDEL (Second panel) green and red channels show GFP-KDEL and DsRed:Talin, respectively. (Third panel) red channel shows DsRed:Talin (G) The tubulin marker protein, TuA:GFP, was expressed with the TGB proteins in TuA:GFP transgenic N. benthamiana. Bar = 20 μm. Photographs were taken at 2 days post infiltration.
In plants infected with PMTV, TGB1 associates with microtubules but microtubule disruption did not have significant effects on PMTV cell-to-cell or systemic movement (Shemyakina et al., 2011; Wright et al., 2010). However, the microtubule network changed as infection spread. The microtubules formed a dense network at the infection front, but after 16 hpi, the network appeared to have a lower density disorganized arrangement. Kinetic experiments over a four day period after agroinfiltration suggests that PMTV TGB1 associations with microtubules precede PD localization (Shemyakina et al., 2011). To determine whether similar effects occurred upon expression of BSMV TGB proteins, TGB1, TGB2 or TGB2/3 derivatives were agroin-filtrated into transgenic N. benthamiana in which green fluorescence is associated with microtubules (TuA-GFP plants). However, in contrast to PMTV, the TGB protein expressing tissue did not exhibit microtubule profile alterations (Fig. 4G). These results thus suggest that transiently expressed BSMV TGB proteins do not elicit substantial microtubule remodeling, in contrast to changes resulting in ER network structures and actin filament patterns.
TGB3 facilitates redistribution of ER vesicles containing actin
To further investigate associations between TGB3 and ER-derived vesicles, we analyzed several TGB3 mutants used in our previous study (Lim et al., 2008). Each derivative was fused to DsRed, and infiltrated into N. benthamiana leaves of plants transformed with the membrane marker GFP-KDEL. After infiltration, GFP-KDEL vesicles containing DsRed:TGB3 were distributed around the nuclei and throughout the cytoplasm (Fig. 5), and upon plasmolysis, a substantial portion of the GFP-KDEL and DsRed:TGB3 fluorescence colocalized as distinct foci at the CW (Fig. 5A), suggesting that the actin-containing ER-derived vesicles also associated with PD desmotubules. The N-terminal TGB3H5L,C11Y and TGB3H5L,C9Y,C11Y mutants, and the central TGB3Q90Y,D91R,L92Y,D93R and TGB3Q115Y,P118R,G120R mutants with amino acid substitutions in the loop between the two membrane spanning domains were all able to bind to TGB2 (Lim et al., 2008). When these four derivatives were infiltrated as DsRed:TGB3 fusions, none of the mutant proteins associated with the CW after plasmolysis and each retracted into the cytoplasm along with the GFP-KDEL fluorescing membranes (Fig. 5B, C, D, and F). Furthermore, the mutants failed to affect the ER network or to induce the actin filament bundling observed with the wtTGB3 protein (data not shown). In contrast, DsRed:TGB3P105R,I108R infiltrated tissue contained very small vesicles distributed near the nucleus and throughout the cell. After plasmolysis, cells expressing the DsRed:TGB3P105R,I108R mutant still had numerous red foci associated with the CW (Fig. 5E), but as is the case with the N-terminal and central-loop TGB3 mutants, actin bundling was not observed (data not shown). We previously showed that the N-terminus of TGB3 is dispensable for PD targeting whereas the C-terminus of the protein is critical for targeting (Lim et al., 2009). In the ER-labeled GFP-KDEL transgenic plants, localization of the TGB316-155 mutant (Lim et al., 2008) lacking the 15 N-terminal TGB3 residues (M-A-M-P-H-P-L-E-C-C-C-P-Q-C-L-) was indistinguishable from wtTGB3 for formation of ER derived vesicles and localization at the PD (data not shown). To test the TGB3 requirements for membrane formation in more detail, the TGB31-150 mutant lacking the five C-terminal residues (-L-S-S-K-R) and a double mutant TGB316-150 mutant lacking both the N- and C-terminal amino acids (Lim et al., 2008) were infiltrated into GFP-KDEL expressing leaves. Both mutants colocalized with ER-derived vesicles showing near the nucleus and within the cytoplasm. Moreover, in agreement with our previous results that TGB3 has a C-terminal PD targeting signal (Lim et al., 2009), plasmolysis experiments revealed that neither of the mutants was able to bind effectively to the CW (Fig. 5G, and data not shown). In sum, our current observations show that the C-terminal deletion derivative is competent to induce ER vesicle formation, but not to associate with actin filaments or to assist in trafficking of vesicles away from the nucleus.

Subcellular localization and effects of TGB3 mutants on ER structure. DsRed:TGB3 mutants were expressed by agroinfiltration into GFP-KDEL transgenic N. benthamiana. Epifluorescent images of DsRed:TGB3 in unplasmolyzed cells (Left) and plasmolyzed cells (Right) were taken at 2 dpi. (A) wt TGB3 (B) TGB3(H5L, C11Y), (C) TGB3(H5L, C9Y, C11Y), (D) TGB3(Q90Y, D91R, L92Y, D93R), (E) TGB3(P105R, I108R), (F) TGB3(Q115Y, P118R, Q120R), (G) TGB3(1–150). Bar = 50 μm.
Latrunculin B interferes with TGB2 and TGB3 actin filament localization
We (Lawrence and Jackson, 2001b) previously reported that cytochalasin D treatment failed to affect TGB1 localization in BSMV infected protoplasts and as a result, postulated that cytoskeletal interactions of the protein were relatively minor. However, the experiments presented above reveal that both actin and the ER undergo major reorganization during BSMV infection and ectopic expression of TGB2 and TGB3. The results further suggested that the subcellular localization of the TGB proteins may be dependent on actin cytoskeleton interactions. Therefore, to reinvestigate actin interactions in more detail, we used Latrunculin B (LatB), which can be up to 100-fold more potent than the cytochalasins, to inhibit actin polymerization in cells infiltrated with TGB derivatives (Baluska et al., 2001; Kim et al., 2005). After LatB treatment, DsRed: Talin patterns in N. benthamiana epidermal leaf cells that were not expressing the TGB proteins exhibited a dramatic shift from a filamentous actin network to disorganized globular structures (Fig. 6A). The fluorescence patterns of GFP:TGB1 expressed alone were not noticeably affected by LatB treatment (Fig. 6B), indicating that TGB1 does not have major associations with the microfilament complex.
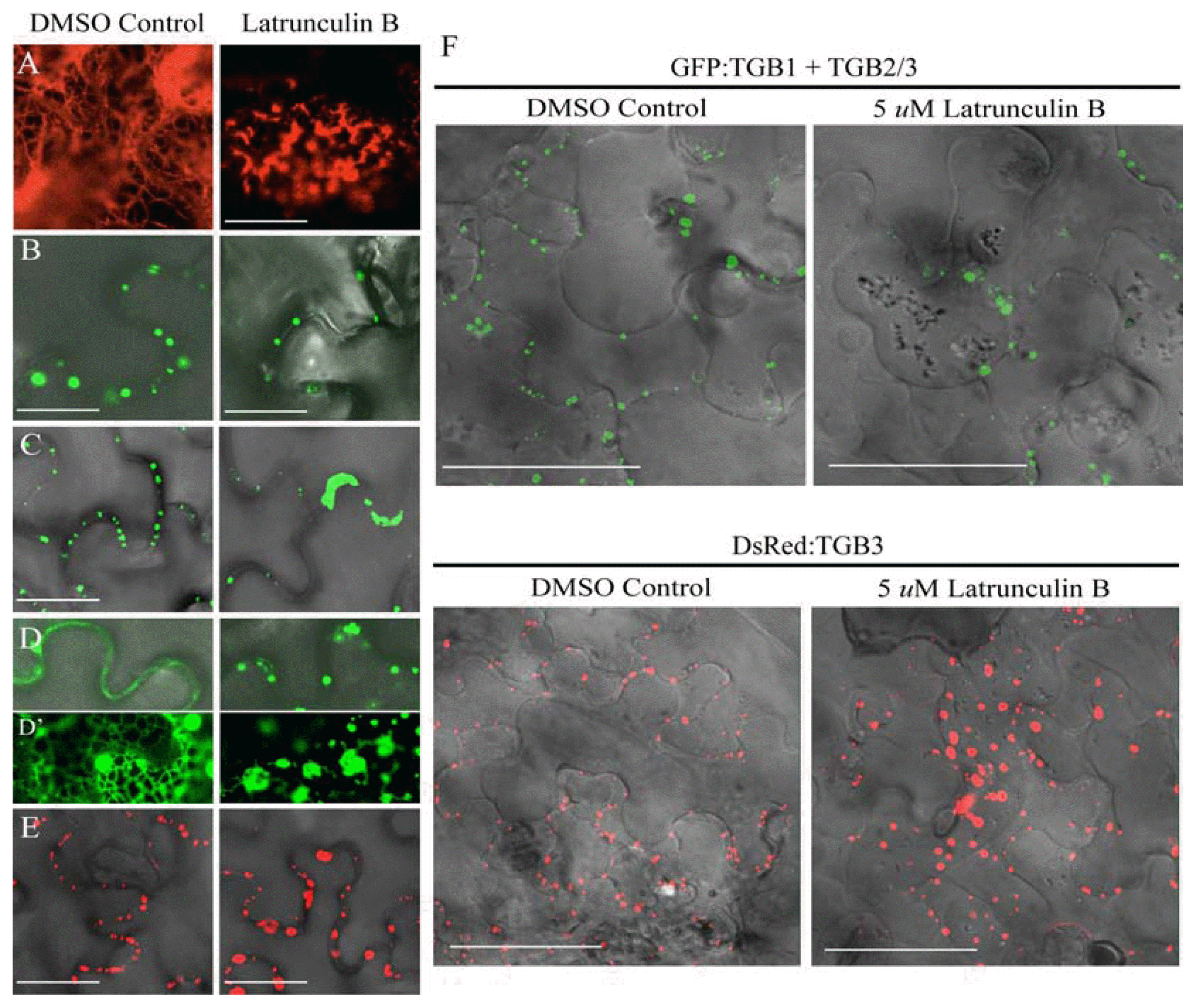
Cytoskeleton disruption by Latrunculin B and TGB localization. The following combinations of proteins were expressed in N. benthamiana epidermal cells by agroinfiltration: (A) DsRed:Talin, (B) GFP:TGB1 (C) GFP:TGB1 + TGB2/3 (D) GFP:TGB2 (E) DsRed:TGB3. The left panels show the localization of the proteins in the absence of Lat B, and the right panels illustrate localization of the proteins after LatB treatment. Bar = 20 μm. The insert panels (D) highlight the network occurring with TGB2. (F) Plasmolysis after LatB treatment. GFP:TGB1 + TGB2/3 and DsRed:TGB3 were expressed in the presence of DMSO (Control), or 5 μM LatB. Both treatments were visualized by confocal microscopy after plasmolysis induced by infiltration with 700 mM sucrose. Bar = 50 μm.
In contrast to the results observed when TGB1 was expressed alone (Fig. 6B), fluorescent foci appeared along adjacent walls of cells co-infiltrated with GFP:TGB1 and the TGB2/3 derivative (Fig. 6C). However, in tissue co-infiltrated with GFP:TGB1 and TGB2/3, the number of CW wall associated foci that typically form in cells (Lim et al., 2009) decreased dramatically compared to those of the DMSO control cells. In this case, fluorescence in aggregated granules in the cytoplasm accounted for most of GFP:TGB1 and only isolated GFP:TGB1 foci were found near the CW of plasmolyzed cells (Fig. 6C). Higher magnification observations of these cells also verified that most of the GFP:TGB1 foci present near or at the CW retracted along with the cytosol after LatB treatment and plasmolysis (Fig. 6F). These results show that when bound to TGB3, TGB1 requires actin cytoskeleton associations to maintain CW attachment.
The patterns of GFP:TGB2, DsRed:TGB3, and GFP: TGB2/3 were each altered after LatB treatment. TGB2 appeared to be generally distributed throughout the cytoplasm when expressed alone, and was converted to heterodisperse fluorescence globular structures in LatB treated cells (Fig. 6D). Higher magnification of the inset panels (Fig. 6D) highlighted the distinctive network appearance that normally occurs when TGB2 is expressed alone and provided better resolution of the disorganization that occurred after LatB treatment. These results suggest that TGB2 and the associated vesicles interact sufficiently with actin to collapse as the cytoskeleton loses its integrity.
Plasmolyzed and unplasmolyzed cells expressing DsRed: TGB3 appeared to form membrane spanning structures along the walls of infiltrated cells (Fig. 6E). However, in striking contrast to the results previously shown with PSLV, in which TGB3 DsRed:TGB3 continued to associate with the cell periphery in LatB treated cells (Schepetilnikov et al., 2008), our results with BSMV revealed that LatB treatment had a major effect on TGB3 targeting (Fig. 6). In tissue not treated with LatB, paired foci were evident along the periphery of the cells and after LatB treatment, the fluorescence mostly appeared in larger globular aggregates (Fig. 6E). The punctate CW foci formed by GFP:TGB1 in the presence of TGB2/3 and DsRed:TGB3, and their dissociation from the CW in the presence of LatB are more evident after plasmolysis (Fig. 6F). These results suggest that actin microfilaments have important roles in the sub-cellular localization of BSMV TGB3 and TGB1 proteins. We have previously shown that TGB3 interacts with both TGB1 and TGB2, and is required for targeting to the CW, so the findings imply that the actin cytoskeleton and, by inference, remodeling of microfilaments, is critical for CW targeting of TGB movement complexes.
Actin filament remodeling has a major role in BSMV cell-to-cell movement
In order to more directly determine the effects of disruption of the actin cytoskeleton on BSMV cell-to-cell movement per se, leaves were divided into three sections on each side of the leaf and the regions on one side were infiltrated with increasing LatB-DMSO concentrations (5 μM, 20 μM, and 50 μM), or the same amounts of DMSO as controls (Fig. 7). Four hours after the LatB-DMSO or DMSO control infiltrations, equal concentrations of BSMV RNA transcripts consisting of wild-type α and β gRNAs and a gRNAγ expressing γb:GFP to assess virus movement (Lawrence and Jackson, 2001a; Lim et al., 2008) were rub-inoculated onto each region of the leaf. At 6 dpi, confocal microscopy revealed that in control regions infiltrated with DMSO alone, BSMV had spread throughout substantial portions of the DMSO infiltrated leaf tissue (Fig. 7). In marked contrast, cell-to-cell movement was progressively reduced in a concentration dependent manner in the LatB treated regions. These results, in conjunction with the TGB subcellular localization experiments, provide persuasive evidence for involvement of a functional actin cytoskeleton in BSMV cell-to-cell movement.

Latrunculin B effects on BSMV movement in N. benthamiana. Leaves were divided into six regions and the left side of the leaf was treated with DMSO dilutions and the right side of the leaves were infiltrated with three different concentrations of LatB (5 μm, 20 μM, or 50 μM) in DMSO. After LatB and DMSO treatments, a BSMV derivative consisting of wild-type α and β RNAs and an RNAg containing the Yb:GFP BSMV was inoculated. GFP visualization of BSMV movement was observed by confocal microscopy at 6 dpi.
Brefeldin A fails to interfere with TGB3 trafficking to the cell wall
Kim et al. (2005) concluded that transport of some intracellular cargo to the plasma membrane (PM) is not actin dependent because LatB treatments failed to affect trafficking from the ER to the Golgi, or from the Golgi to the PM. A more direct assay to interfere with post-Golgi vesicle transport to the PM is inhibition by BFA (Nebenfuhr et al., 2002). To investigate whether TGB intracellular targeting depended on movement through the Golgi, we co-expressed the ST-GFP Golgi marker in N. benthamiana, along with DsRed:TGB3 or DsRed:TGB3P105R,I108R, which disrupts TGB3-TGB2 interactions. After agroinfiltration, the ST-GFP Golgi marker was detected at the cell periphery and the fluorescent vesicles retracted with the plasma membrane after plasmolysis (Fig. 8A). BFA treatments also resulted in a dramatic disorientation of ST-GFP membranes, indicating that the Golgi integrity was affected by the inhibitor. Although the DsRed:TGB3 protein appeared to localize with the Golgi in unplasmolyzed cells, it remained associated with the CW after plasmolysis, with or without BFA treatment, indicating that transit to the CW can occur independently of a functional Golgi apparatus (Fig. 8B).
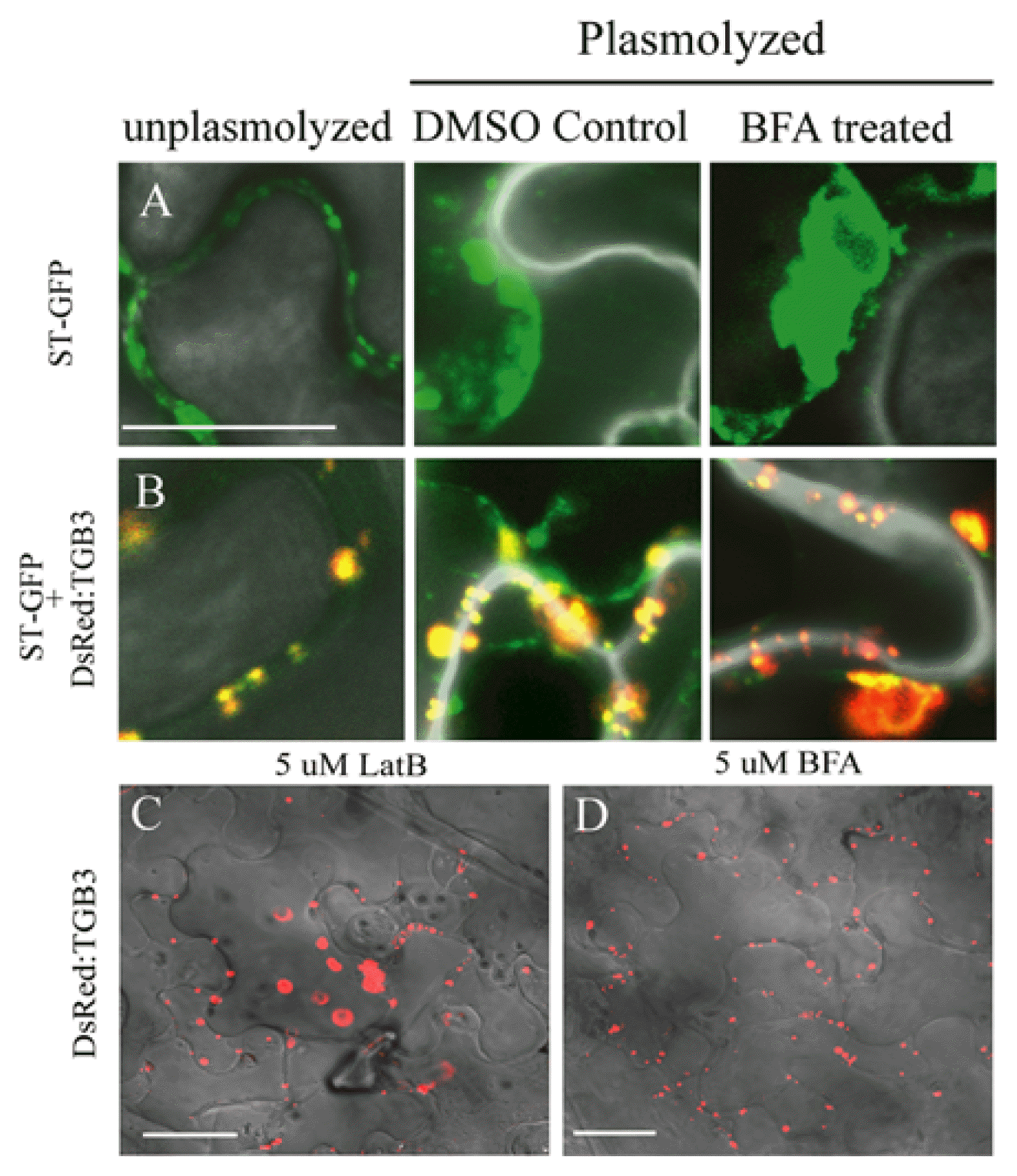
Brefeldin A disruption of Golgi and effects on DsRed: TGB3 localization. The DsRed fusions to wtTGB3 derivatives were coexpressed with the ST:GFP Golgi marker protein in N. benthamiana epidermal cells. Cells were infiltrated with DMSO in buffer, or were treated with BFA to disrupt Golgi. Labels at the top of the columns designate whether or not cells were plasmolyzed. (A) ST-GFP, (B) DsRed:TGB3 plus ST-GFP, Bar = 20 μm. (C) Comparison of DsRed:TGB3 expression in LatB or (D) BFA treated wild type N. benthamiana after plasmolysis. Bar = 50 μm. Fluorescent images were captured at 2–3 dpi and overlayed onto DIC images.
A direct comparison of differences in the requirements of microfilaments and the Golgi for TGB3 CW targeting is shown in Fig. 8C and 8D. These images clearly demonstrate that after plasmolysis, actin disruption results in retraction of TGB3 from the CW (Fig. 8C), and, that TGB3 remains associated with the CW after Golgi disruption in BFA treated tissue (Fig. 8D). Interestingly, the BSMV TGB3 BFA results are in agreement with similar experiments with PLSV TGB3 in which BFA treatments failed to affect association with “peripheral bodies” (Schepetilnikov et al., 2008). In marked contrast, our actin inhibition experiments provide different results from those with PSLV TGB3 (Schepetilnikov et al., 2008) because BSMV TGB3 failed to localize to the CW in LatB (Fig. 8C) treated tissue, whereas PSLV TGB3 “peripheral body” localization was not dramatically affected by LatB treatment.
Discussion
We have previously recovered BSMV TGB1 ribonucleo-protein complexes that are thought to function during cell-to-cell movement in infected plants, and have shown that the abundance and ratios of BSMV TGB proteins regulate both intracellular and intercellular movement (Lim et al., 2008). The BSMV GFP:TGB1 reporter derivative formed punctate foci at the surface of protoplasts and also localized at the CW of infected plant cells (Lawrence and Jackson, 2001b; Lim et al., 2009). In the absence of TGB3, ectopically expressed TGB1 colocalizes with perinuclear vesicles during BSMV infections of protoplasts and whole plants. Studies with other virgaviruses have shown that small amounts of TGB1 localize to the nucleolar region during ectopic expression of PSLV and PMTV, and that nucleolar targeting signals are found in TGB1 N-terminal motifs involved in RNA binding and/or long distance movement (Semashko et al., 2012a, b; Torrance et al., 2011; Wright et al., 2010). The significance of these observations for cell-to-cell and vascular movement is unclear, but the results do add to a long list of multifunctional activities of “hordei-like” TGB1 proteins.
A role for TGB3 in subcellular localization of TGB complexes was first noted in protoplasts infected with BSMV mutants that were compromised in expression of TGB3. Plants infected with these mutants exhibited a dramatic shift of GFP:TGB1 from the plasma membrane to perinuclear membranes (Lawrence and Jackson, 2001b), and similar results have been observed in leaf cells (Lim et al., 2009). Several experiments also indicate that TGB2 associates with the ER network and interacts directly with TGB3 to mediate PD targeting (Lim et al., 2008; Lim et al., 2009). These results have lead to a model positing that TGB3 functions in vivo by forming a bridge between TGB1 and TGB2 by binding directly to both proteins, and facilitates targeting of the resulting TGB complex to the PD. The biological importance of these interactions has been demonstrated by the failure of BSMV to move from cell-to-cell in leaves inoculated with mutants containing amino acid substitutions that disrupt TGB1/TGB3 or TGB2/TGB3 interactions (Lim et al., 2008; Lim et al., 2009). Clearly, the TGB proteins elicit major cytological alterations that are required for cell-to-cell movement, and such interactions provide a framework for more extensive mechanistic studies to unravel details of BSMV movement.
Our current results verify and extend earlier findings related to BSMV movement (Jackson et al., 2009). We have now shown that BSMV infection elicits actin remodeling at the infection front to produce filament thickening, and have provided evidence that the actin remodeling may facilitate subcellular targeting of the TGB proteins and cell-to-cell movement of BSMV. Agrobacterium ectopic expression experiments to dissect TGB functions demonstrate that TGB1 has little effect on actin filament patterns, but that TGB3 elicits formation of thick actin cables similar to those appearing during infection, whereas intermediate filaments form in cells expressing TGB2. Moreover, LatB infiltration experiments disrupting actin filament integrity indicate that a functional cytoskeleton is essential for PD localization of TGB protein complexes and for optimal BSMV cell-to-cell movement. Hence, these results show that in addition to its requirement for PD localization of TGB1 and TGB2, BSMV TGB3 functions in host interactions that culminate in cytoskeleton remodeling.
As well as cytoskeleton remodelling, BSMV infection results in an increased abundance of membrane vesicles in GFP-KDEL transgenic N. benthamiana. Ectopic expression of BSMV TGB1 had little obvious effect on N. benthamiana membrane structure and, aside from eliciting a more defined network, TGB2 expression appears to have minimal effects on membrane structure. TGB1 and TGB2 also localized with membrane vesicles even though extensive membrane proliferation was not observed. However, ectopically expressed TGB3 elicited formation of a complex and well-defined ER network that is closely associated with, or houses, thick actin cables and TGB proteins. TGB3 also appeared to function in redistribution of membrane vesicles throughout the cytosol. Virus movement and membrane reorganization, which is especially obvious in the perinuclear region, was disrupted by mutations in the central membrane-spanning domain of TGB3. Our findings thus suggest that host membrane associations are involved in several aspects of BSMV movement that merit future study.
In addition to differences in requirements of the coat protein for cell-to-cell movement of hordeiviruses and potexviruses, a number of variations are evident in TGB1 protein structure, biochemical activities, interference with host gene silencing, and movement functions of the virga-viruses and the TGB-containing flexiviruses (Jackson et al., 2009; Morozov and Solovyev, 2003; Verchot et al., 2010). A recent paper illuminating a TGB1 requirement for formation of X bodies associated with PVX highlights another major difference in functions of the TGB1 proteins of the hordeiviruses and the potexviruses that relates to BSMV actin remodeling. In contrast to BSMV, in which TGB3 expression has a major effect on actin architecture, PVX TGB1 is essential for X-body formation and functions in extensive actin and membrane remodeling (Tilsner et al., 2012). The multilayered membranous X-body is an important organelle that is required for normal levels of viral RNA replication and virion accumulation. Nevertheless, in plants and protoplasts infected with PVX mutants unable to express TGB1, morphogenesis of X bodies fails to occur, yet low levels of PVX replication can be detected and small amounts of virus particles accumulate (Tilsner et al., 2012). However, ectopic expression of PVX TGB1 results in massive remodeling of host actin and endomembranes, and recruitment of these structures, as well as TGB2 and TGB3, to sites near the nucleus. Subsequently, X-bodies develop into complex multilayered membrane organelles adjacent to the nucleus, that selectively incorporate TGB proteins, ribosomes, viral RNA and virions to specific sites within the granular vesicular bodies (Tilsner et al., 2012). These differences between the two viruses are further illustrated by conventional electron microscopic observations showing that structures corresponding to PVX X-bodies are not present in BSMV-infected cells, and that BSMV replicates in membrane vesicles formed from the chloroplast outer membrane (Lin and Langenberg, 1985).
Our current results add to a growing list of major differences in the movement processes of TGB-encoding viruses (Verchot et al., 2010). We (Lawrence and Jackson, 2001b) previously reported that cytochalasin D treatment failed to affect TGB1 localization in BSMV infected protoplasts and as a result, postulated that cytoskeletal interactions of the protein were relatively minor. However, the experiments presented here reveal both actin remodeling and changes to ER structure as a consequence of BSMV infection and transient expression of TGB3 and TGB2/3. Our observations also provide evidence that the subcellular localization of the TGB proteins depends on actin cytoskeleton interactions. To investigate these interactions in more detail, we used LatB to inhibit actin polymerization (Baluska et al., 2001; Kim et al., 2005) in cells infiltrated with TGB reporter proteins. In contrast to cytochalasin D used in our earlier experiments (Lawrence and Jackson, 2001b), LatB can be up to 100-fold more potent than cytochalasins, and functions by shortening and thickening of actin filaments. After LatB treatment, the DsRed:Talin patterns in N. benthamiana infiltrated epidermal leaf cells exhibited a major shift from a filamentous actin network to thick cable-like structures. LatB inhibition also reduced BSMV movement and disrupted TGB targeting to the PD. From these experiments, we conclude that actin cytoskeleton modifications are required for BSMV movement and that TGB3 has a critical role in cytoskeleton remodeling during movement.
In contrast to the BSMV LatB experiments described above, experiments with the closely related PSLV TGB3 have resulted in different conclusions about mechanisms functioning in PD targeting (Schepetilnikov et al., 2008). In the case of BSMV, actin cytoskeleton disruption by LatB interfered with CW localization of TGB3, and TGB1 when coexpressed with TGB2/3, whereas PSLV TGB3 CW localization was not dramatically affected by LatB treatment (Schepetilnikov et al., 2008). These disparate results highlight fundamental differences in the mechanisms of sub-cellular transit of BSMV and PSLV. Such differences between related viruses may occur more often than previously realized, as illustrated by a previous report describing differences in the movement of two tobamoviruses (Harries et al., 2009). In this direct comparison, movement of TMV is strongly inhibited by LatB treatment, whereas movement of the related TVCV is unaffected by LatB treatment. These results argue strongly that more than one mechanism may be operative in some closely related viruses, and our collective results suggest that BSMV and PSLV may fit within this category.
Evidence for TGB3 associations with the Golgi membranes during coexpression of DsRed:TGB3 and the ST-GFP Golgi marker indicates that Golgi derived vesicles and DsRed:TGB3 co-localize with the CW after plasmolysis. BFA interference with Golgi stack integrity resulted in a major collapse of vesicles localized in close proximity to the CW, but BFA appears to have only limited effects on BSMV localization or PSLV TGB3 associations with “peripheral bodies” (Schepetilnikov et al., 2008). Nevertheless, differences in the BSMV and PSLV LatB cytoskeleton disruption experiments suggest that different mechanisms may function in some TGB3 interactions culminating in PD targeting.
Other than the preliminary experiments shown above, which suggest that BSMV infection does not result in obvious changes to microtubules, we have not extensively investigated possible direct interactions of BSMV TGB proteins with microtubules. However, in other experiments with the related Potato mop-top virus (PMTV), colchicine treatments were used to disrupt tubulin polymerization and microtubule integrity (Shemyakina et al., 2011; Wright et al., 2010). Colchicine can affect multiple metabolic and regulatory processes affecting a large number of functions that might interfere with TGB1 localization to the CW. However, the PD associations of PMTV mutants provided evidence for an association between microtubules and PMTV TGB1. Of particular interest, cells were observed for several days after transient expression of the three TGB proteins in ratios corresponding to those occurring during virus infection. During this period, a defined series of kinetic events were noted, beginning with PMTV TGB1 nucleolar interactions and proceeding through cytoplasmic granules to the CW. Thus, the effects of BSMV and PMTV on microtubule remodeling, seem to differ, and these experiments reinforce our suggestion that multiple pathways may operate in CW targeting during TGB1 expression of the virgaviruses.
Unfortunately, individual events involved in viral movement from subcellular sites of replication to the PD and adjacent cells are difficult to dissect experimentally, and many of these problems have been discussed previously (Harries et al., 2010). The infection front where important events are coordinated is a moving boundary consisting of a limited number of cells undergoing a series of asynchronous steps, so relatively few studies have probed events at this stage of infection. Variations in delivery protocols also contribute to experimental differences or artifacts that can lead to aberrant subcellular trafficking effects. In this regard, examples of the effects of overexpression of PSLV TGB3 has been described recently in which anomalous cell death, membrane abnormalities and disrupted Golgi functions occur during transient infection (Solovyev et al., 2012). Third, pharmacological approaches can be quite variable in the hands of different researchers. Finally, a more diverse array of approaches, including infectivity studies applied to different hosts might provide interesting insights into alternative strategies employed by BSMV and other hordei-viruses. Although it would be preferable to investigate movement in the natural BSMV cereal hosts, these plants present technical difficulties that are difficult to circumvent. Fortunately, BSMV, unlike PSLV, is able to infect N. benthamiana, so we have been able to compare cytological and biochemical experiments with infectivity results in this host.
Acknowledgments
We thank Drs. Steven Ruzin and Denise Schichnes for advice and support with microscopy carried out in conjunction with the research, and to the Biological Imaging Facility at University of California-Berkeley for access to the Zeiss LSM 510 confocal microscope used during the research. We also thank Nam-Hai Chua for the DsRed-talin plasmid, Valarian Dolja for the ST-GFP marker, and David Baulcombe for transgenic N. benthamiana seeds expressing GFP-KDEL and TuA-GFP. This work was supported by grants from the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center No. PJ 0090332012), Rural Development Administration, and the National Research Foundation of Korea (NRF No. 2012R1A1A2007417), and by U.S. Department of Agriculture Competitive Research Grant 2008-35319-19225 to AOJ.