Bacterial Traits Involved in Colonization of Arabidopsis thaliana Roots by Bacillus amyloliquefaciens FZB42
Article information
Abstract
Colonization studies previously performed with a green-fluorescent-protein, GFP, labeled derivative of Bacillus amyloliquefaciens FZB42 revealed that the bacterium behaved different in colonizing surfaces of plant roots of different species (Fan et al., 2012). In order to extend these studies and to elucidate which genes are crucial for root colonization, we applied targeted mutant strains to Arabidopsis seedlings. The fates of root colonization in mutant strains impaired in synthesis of alternative sigma factors, non-ribosomal synthesis of lipopeptides and polyketides, biofilm formation, swarming motility, and plant growth promoting activity were analyzed by confocal laser scanning microscopy. Whilst the wild-type strain heavily colonized surfaces of root tips and lateral roots, the mutant strains were impaired in their ability to colonize root tips and most of them were unable to colonize lateral roots. Ability to colonize plant roots is not only dependent on the ability to form biofilms or swarming motility. Six mutants, deficient in abrB-, sigH-, sigD-, nrfA-, yusV and RBAM017410, but not affected in biofilm formation, displayed significantly reduced root colonization. The nrfA- and yusV-mutant strains colonized border cells and, partly, root surfaces but did not colonize root tips or lateral roots.
Plant growth-promoting rhizobacteria, PGPR, is a diverse group of bacteria able to colonize plant roots and to act beneficial on their growth (Lugtenberg and Kamilova, 2009). They contribute to increased yield of cereals, vegetables and other plants of economical importance (Fan et al., 2011). Mainly, plant-associated representatives of the genus Bacillus are commercialized successfully as biofertilizer and biological control agents in agriculture. An important property of those bacteria is their ability to form thermostable and chemically resistant endospores, allowing preparing stable long-lasting products (Borriss, 2011). Competitive rhizosphere colonization is crucial for plant-PGPR interactions (Chin-A-Woeng et al., 2000), but in contrast to Gram-negative PGPR, little is known about colonization pattern of Gram-positive PGPR.
The type strain for the group of plant-associated bacilli, recently classified as Bacillus amyloliquefaciens subsp. plantarum (Borriss et al., 2011), is B. amyloliquefaciens FZB42. Its genome is completely sequenced and mapped and several genes responsible for its plant growth promoting activity are uncovered (Chen et al., 2007; Idriss et al., 2002; Idris et al., 2007). The genome analysis of FZB42 revealed numerous gene clusters coding for the synthesis of non-ribosomal cyclic lipopeptides and polyketides with antibacterial and antifungal action. Mutant strains impaired in its capability to synthesize those compounds were generated by site-directed cassette mutagenesis via homologous recombination (Chen et al., 2009a; Chen et al., 2009b). Moreover, a screen performed with a mariner transposon TnYLB-1 library prepared from FZB42, revealed three genes important for plant growth promotion (Budiharjo, 2011).
Colonization studies previously performed with a green-fluorescent-protein, GFP, labeled derivative of B. amyloliquefaciens FZB42 suggested that the bacterium behaves different in colonizing surfaces of plant roots of different species. In contrast to maize, FZB42 colonized preferentially root tips when colonizing Arabidopsis thaliana. On Lemna minor, FZB42 accumulated preferably along the grooves between epidermal cells of roots and in the concave spaces on ventral sides of fronds (Fan et al., 2011; Fan et al., 2012).
In order to extend these studies and to elucidate which genes are crucial for root colonization, we used here derivatives from GFP labeled B. amyloliquefaciens FZB42 containing targeted mutations for studying the fate of colonizing the roots of Arabidopsis seedlings. The fate of root colonization in mutant strains impaired in synthesis of alternative sigma-factors, non-ribosomal synthesis of lipopeptides and polyketides, motility, biofilm formation, and plant-growth-promoting activity was analyzed by confocal laser scanning microscopy and compared with the pattern of the FZB42 wild type strain. Due to autofluorescence of soil particles, the study was performed in a gnotobiotic system using Arabidopsis seedlings grown in agar solidified Murashige-Skoog medium. It has been already shown that results obtained with FZB42 grown in gnotobiotic systems and soil systems are comparable (Fan et al., 2012).
Material and Methods
Bacterial strains and growth conditions
The bacterial strains, used in this study, are listed in Table 1. The bacteria were cultivated routinely on Luria broth (LB) medium solidified with 1.5% agar. Biofilm formation was followed in MSgg medium (Branda et al., 2001) without shaking for four days at room temperature. Fermentation in liquid media (50 ml) was carried out in 500 ml flasks at 37°C and 200 rpm. The media and buffer used for DNA transformation of Bacillus cells were prepared as previously described (Koumoutsi et al., 2004).
Construction of mutant strains
To generate a transposon library of FZB42 we followed the procedure described by Haldenwang (Le Breton et al., 2006). Plasmid pMarA carrying the mariner based transposon TnYLB-1 was transformed into B. amyloliquefaciens FZB42. Transformants were screened for plasmid-associated properties, i.e. kanamycin resistance at a permissive temperature for plasmid replication (30°C) and kanamycin resistance at the restrictive temperature (48°C). Transformants were verified by digesting with restriction endonucleases. Plasmid-containing colonies were incubated overnight in LB medium at 37°C. Then samples were plated out on LB agar containing kanamycin and incubated at 48°C to select for transposants. Mutant strains carrying the gfp-plus gene were constructed from the knock-out mutants listed in Table 1 via homologous recombination with FB01mut. Chromosomal DNA isolated from the mutant strains was introduced into competent FB01mut cells. To construct the gfp-labeled mutant bearing a knock-out mutation of the pznA-gene the plasmid pRS32b was transformed into competent cells of FB01mut. All mutant strains were checked for the appropriate genes by colony PCR using gene specific primers. To verify that all transformants still carry the GFP they were observed with a Zeiss SteneSU6 binocular with exelement-citation filter of 450–490 nm and a LP520 emission filter.
Sterilization of Arabidopsis seeds
For plant growth promotion and colonization studies, the A. thaliana ecotype Columbia-0 was used. The seeds were surface sterilized with 10% (NaClO) over a period of 3 minutes. To remove the bleach, sterile distilled water was added, and changed four times. The sterile seeds were transferred onto plates containing Murashige and Skoog (MS) medium (50%) with 0.6% agar and 1% sucrose and incubated at 22°C under long day light conditions (16 h light/8 h dark) for seven days.
Assay for plant growth promotion with A. thaliana
The roots of seven-day-old Arabidopsis seedlings were dipped into the bacterial spore suspension (1 × 105 cfu/ml) for 5 min and four seedlings were transferred into a square Petri dish containing half-strength MS medium solidified with 1% agar. The square Petri dishes were incubated in a growth chamber at 22°C and a daily photoperiod of 16 h. Fresh weight of the plants was measured 21 days after transplanting.
Monitoring of bacteria on plant roots and statistical analysis
Bacteria from fresh mutant colonies were grown in Luria broth medium without antibiotics up to an OD600 1.0 and then diluted 104 fold yielding 1 × 105 cfu/ml. The seven-days old seedlings were taken from the plates and their roots were incubated with the spore suspension for two minutes. Afterwards, they were transferred onto square Petri dishes (12 × 12 cm) containing 50% MS and 0.8% agar but without sucrose. The square plates were kept in a vertical position during the incubation time of seven days at 22°C under long day light conditions (16 h light/8 h darkness) in a plant growth chamber.
After growing for seven days the inoculated plant seedlings were rinsed in sterile saline for three times by dipping the whole seedling into a drop of one ml saline for 10 times. Than the seedling was vortexed for 5 seconds in an Eppendorf tube containing 1 ml sterile saline and transferred into another tube with 1 ml saline and vortexed for 20 seconds. Between every washing step the seedling was dipped on a dry, sterile Petri-dish to remove the remaining saline. Than the suspension was diluted two times 1:20 with saline (50 3l: 950 3l). From both dilutions (1:20, 1:400) 100 3l were plated on selective plates. They were incubated over night at 37°C and the colonies were counted the next day. This procedure was performed with three individual plants per experiment. Per experiment five whole plants and their leaves were weighted to calculate the root fresh weight. This was used to determine the cfu per standardized unit [cfu/mg root fresh weight].
This experiment was repeated eight times. In order to determine the level of colonization for each strain the weighted mean and the variance was calculated with the following formulas:
For two strains one experiment each was rejected respectively because all three plants had zero cfu and therefore an average and variance of zero (Division by zero is forbidden). One strain was arbitrarily set to zero with no variance because in 5 of eight experiences all three plants had no cfu. In order to estimate the statistical significance a student’s t-test was performed with the whole set of data. For calculation “Microsoft Office Exel 2003” was employed. All knock out mutants were compared with FB01mut1 to detect differences in root colonization.
Results
Construction of fluorescence protein-labeled mutant strains
We introduced the mariner-based transposon TnYLB-1 by DNA-mediated transformation into competent cells of FZB42. We isolated randomly several mutants, impaired in their ability to colonize plant surfaces and to promote plant growth, using specific screening procedures which will be described elsewhere (Chowdhury et al., in preparation, Budiharjo, 2011). Several mutants impaired in swarming (degU), biofilm formation (pabA, yusV), and plant growth promotion (RBAM017410, abrB, nfrA) were isolated. Chromosomal DNA isolated from those mutant strains was used to transform competent cells of strain FB01mut expressing green fluorescent protein, GFP (Fan et al., 2011). In the same way also GFP-labelled strains, deficient in the alternative sigma factors SigH and SigD, nonribosomal synthesis of lipopeptides and polyketides, and in synthesizing plantazolicin (Scholz et al., 2011), were constructed. All transformants were checked for their ability to express GFP (see Methods). The GFP-labelled constructs listed in Table 1 were used for further root colonization studies.
Biofilm formation
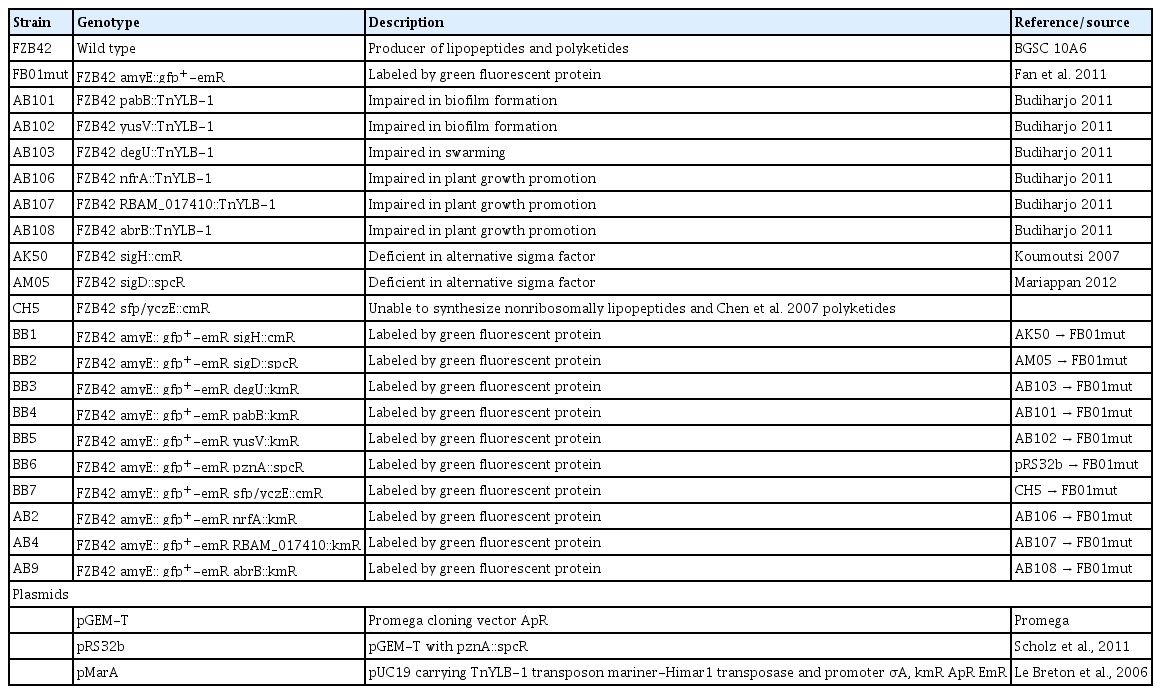
Biofilm formation is a precondition for colonizing plant roots and exerting beneficial effects on plant growth. Therefore, FB01mut and the mutant strains impaired in several cellular functions were checked for their ability to form biofilms in liquid MSgg medium (see Methods). FZB42 wild-type and FB01mut developed well aerial structures of whitish color. They appeared to be dry and of cotton like texture. The previously colorless MSgg medium became brownish. Fig. 1 shows biofilm formation of the original FZB42 mutant strains (lower sets) and their transformed GFP-labeled counter-parts. All pairs showed a similar ability to form biofilms indicating that that the presence of GFP does not affect the quality of biofilms. Reduced biofilm formation was registered for mutant strains deficient in the sfp, pabB and degU genes. The ΔpabB mutant only displayed an unstructured layer of surface-attached cells. The medium did not change to brownish color. More surface structures were built by CH5 (sfp/yczE mutant), but they were still tiny and appeared slimy. Also the ΔdegU-mutant produced tiny structures; they appeared to swim on the medium as isolated islands. The ΔpznA mutant exhibited a slightly reduced biofilm formation suggesting that plantazolicin might affect in some way biofilm formation. There was no apparent difference to the wild type in the other mutant strains deficient in nrfA, abrB, RBAM017410, yusV, sigD, and sigH.
Biofilm formation of mutant strains: The upper set represents the biofilm formation of the FZB42 mutant strains and the lower set their GFP expressing counterparts. (A) nfrA-mutant: normal biofilm formation; (B) abrB-mutant: normal biofilm formation; (C) pabB-mutant: no biofilm, but few surface attached cells; (D) sfp/yczE-mutant: reduced biofilm formation with slimy surface; (E) RBAM017410-mutant: normal biofilm formation; (F) degU-mutant: reduced and altered biofilm formation; (G) pznA-mutant: reduced biofilm formation; (H) yusV-mutant: normal biofilm formation; (I) sigH-mutant: normal biofilm formation; (J) sigD-mutant: normal biofilm formation.
Monitoring of root colonization
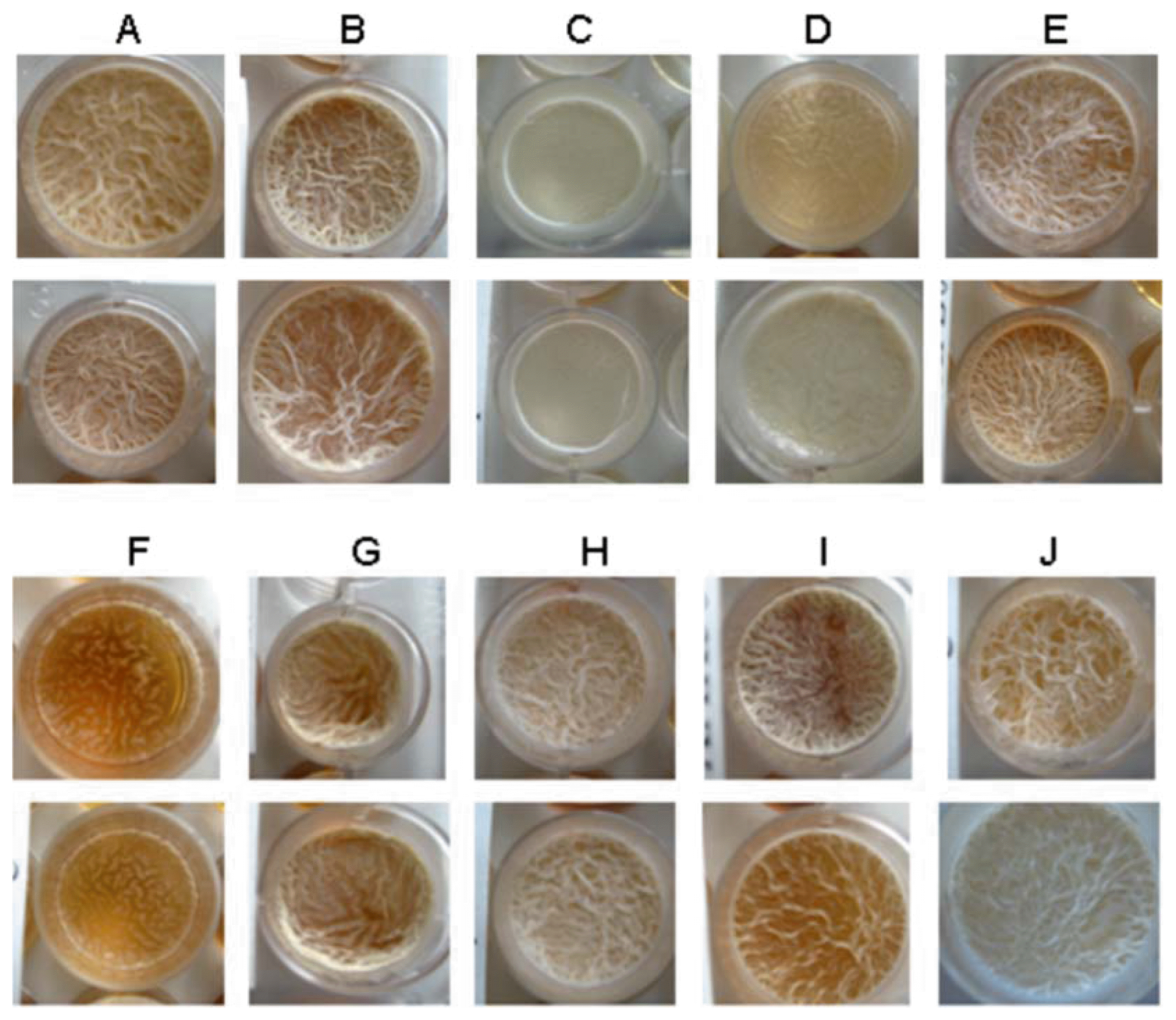
Colonization studies were performed in a gnotobiotic system in which the bacteria were added to seven day-old seedlings of A. thaliana (see Methods). Despite a high variability in eight independent experiments, it became evident that all mutant strains have a decreased ability to colonize plant roots compared to the wild type. The sigH mutant strain was completely unable to colonize plant roots suggesting that genes under control of SigH are indispensable for root colonization by FZB42. Very weak colonization rates were observed for mutant strains deficient in sfp/yczE, abrB, RBAM017410, and degU. shows the colonization rate for each strain. The highest colonization rate was found in wild-type strain FB01mut1 with a mean of more than 16,000 cfu/mg root fresh weight. By contrast, FB01mut1xAK50 (ΔsigH) showed no ability to colonize the Arabidopsis roots. Although, there are large deviations in sigH-, sfp/yczE- abrB-, RBAM017410- and degU-mutants, a very weak colonization rate is clearly observed. The nrfA-mutant showed a reduced colonization rate of approximately half the amount of cfu/mg compared with FB01mut1 with a significance of 92%. However, the nrfA-mutant data are statistically not significant. For the other strains (yusV-, pabB- and pznA-mutants) no statement can be made due to a lack of statistical significance.
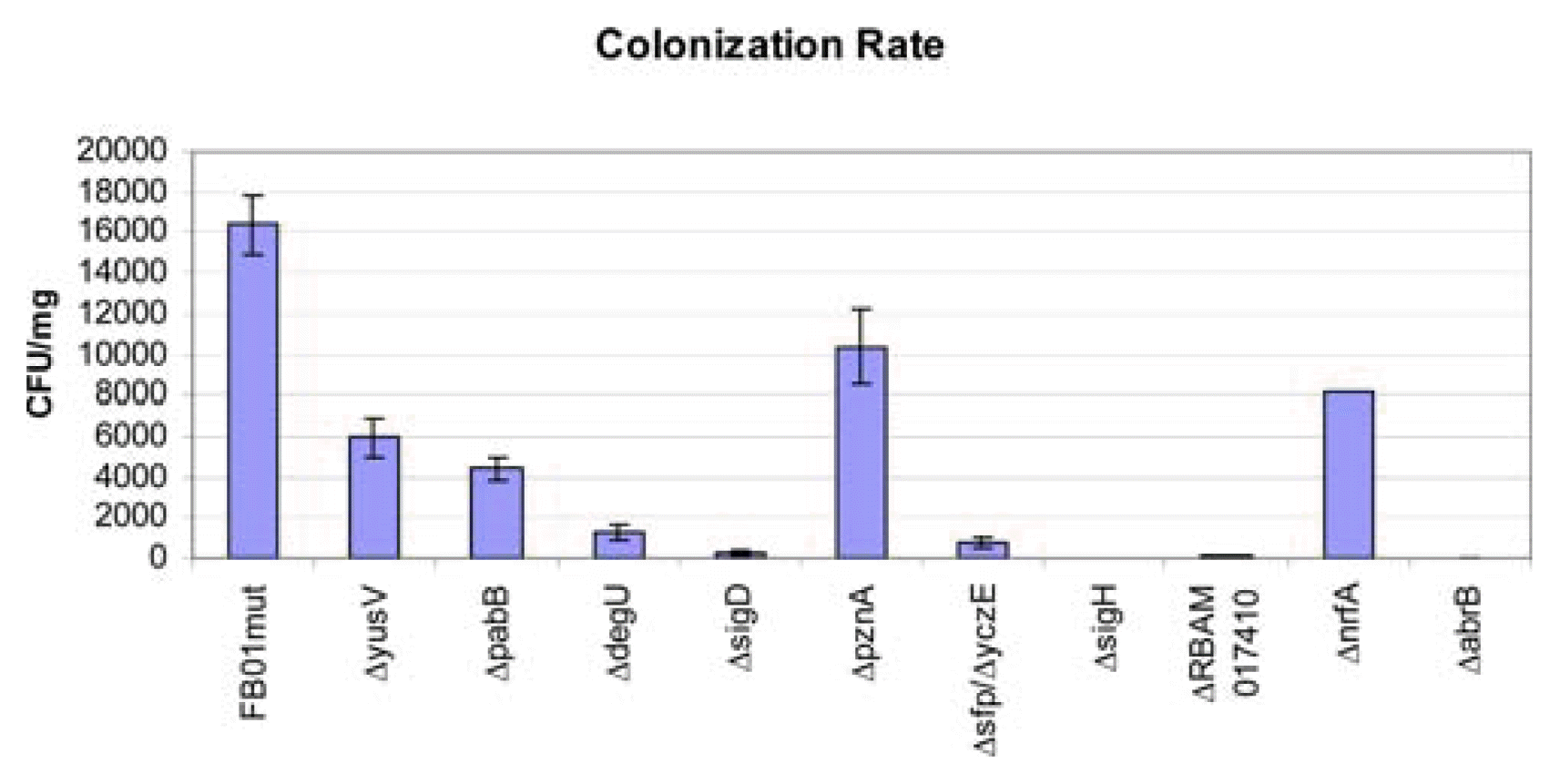
The fate of root colonization was followed by confocal laser scanning microscopy, CLSM, of Arabidopsis seedling roots taken seven days after inoculation with the mutant strains. Strain FB01 could colonize the whole root surface including the root tip and the border cells (Fig. 2A) as well as the growing lateral roots (Fig. 2B). FB01mut colonized especially the junction between epidermal cells and root hairs. Massive colonization of Arabidopsis root hairs and main root tips by FZB42 has been previously reported. It is assumed that these locations are sites of intensive root exudation which is attracting the bacteria (Fan et al., 2012).
Root colonization rate of wild-type (FB01) and mutant strains. Seven days after inoculation the number of colony forming units (CFU) per mg root fresh weight was determined (see methods).
Notably, all GFP labeled mutant strains exhibited a reduced capability to colonize Arabidopsis roots, but interesting differences in the fate and intensity of root colonization were observed. The ΔpaB-mutant did not colonize the root tip, but was found in surrounding border cells (Fig. 2C). Several clusters of fluorescent bacteria were found along the main root surface while the lateral root was not colonized (Fig. 2D). Similarly, the yusV mutant strain did also not colonize the root tips, but some bacteria were growing at border cells forming the root cap together with the mucilage (Fig. 2E) which seemed to be a very nutrient rich area in which plant defense is not as efficient as directly at the root surface. This behavior indicates a reduced capability to overcome plant defense reactions in this mutant strain. There were almost no bacteria present at the lateral root as well as on the root surface. However, opposite to the lateral root there was a cluster of bacteria possibly originating from some colonized border cells, which became detached during root preparation (Fig. 2F). The degU mutant strain was found unable to colonize Arabidopsis roots and grew rarely at some border cells. It did not colonize the root tip, the root surface or the lateral root (Fig. 2G and Fig. 2H). Similarly, no traces of root colonization were detected in ΔsigD, Δsfp/yczE, and ΔsigH (not shown).
By contrast, the pznA mutant grew along on the root surface in vicinity of the root tip and on some border cells but not directly on the root tip (Fig. 2I). The base of the lateral root was also colonized by the pnzA mutant strain (Fig. 2J) indicating that ability to colonize plant roots is relatively less impaired in the pnzA mutant. Colonizing roots was more impaired in the nrfA mutant although some patches of fluorescent cells were detected at border cells (Fig. 2K) and at the main root surface (Fig. 2L). Almost no root colonization was detected in mutant ΔRBAM01740 except some fluorescent cells growing in vicinity of the root cap (Fig. 2M and Fig. 2N). The same was true for mutant abrB but there were clearly fluorescent cells growing on border cells of the root cap (Fig. 2O).
Discussion
Ten GFP-labeled mutant strains of the plant associated B. amyloliquefaciens FZB42 was analyzed for their ability to colonize the roots of A. thaliana seedlings. Mutants impaired in swarming motility, biofilm formation, and plant growth promotion were identified after transposon mutagenesis of FZB42 (Budiharjo, 2011). In order to find out whether those mutant strains were also affected in their ability to colonize plant roots, we labeled the mutant strains by GFP and followed their fate of colonizing of plant roots by confocal laser scanning microscopy. In addition, several targeted mutants impaired in alternative sigma factors, nonribosomal synthesis of secondary metabolites, including surfactin, and in synthesis of plantazolicin was GFP labeled and were also included in this study. For technical reasons, only total root colonization rates including statistical analysis were detetermined. It ruled out that especially strains with knock out mutations in alternative sigma factors, abrB, and the unknown gene RBAM017410 were heavily affected in their ability to colonize plant roots (Fig. 2). Interestingly, mutant strains were distinguished from wild type by their inability to colonize Arabidopsis root tips, whilst border cells were colonized by most mutant strains. The results are summarized in Table 2. The strains carrying knock-out mutations in the sfp, pabB-, pznA-, and in the degU genes were simultaneously affected in their ability to form biofilm and/or swarming motility and to colonize plant roots. That is not surprising, since swarming and biofilm formation are thought as being necessary preconditions for surface colonization. On the other hand, it ruled out that the ability to colonize plant roots is not only dependent on the ability to form biofilms or on swarming motility. Six mutants, deficient in abrB-, sigH-, sigD-, nrfA-, yusV and RBAM017410, displayed significantly reduced root colonization but were not affected in biofilm formation. The nrfA- and yusV-mutant strains colonized border cells and, partly, root surfaces but did not colonize root tips or lateral roots.
Except the mutant harboring a gene defect in the pabB gene, whose growth is dependent on presence of para-amino-benzoic acid, we did not found any differences in growth behavior of wt and mutant strains under laboratory conditions. However, as stated in Table 2, many mutants used in this study had an apparent phenotype when growing on plant roots. Mutant strains yusV, sfp/yczE and pabB were found impaired in biofilm formation, whilst degU and sigD were affected in swarming motility, for example. At recent, we can’t exclude that the reason for reduced colonization in other mutant strains is due to some growth deficiencies which are not directly linked with plant colonization.
Our results point out that ability to colonize plant roots is a “conditio sine qua non” for plant growth promotion suggesting that direct contact between bacteria and plants is necessary. Especially, the reduced ability to grow at plant root surfaces, which we observed in the mutant strains impaired in plant growth promotion, abrB, RBAM01704, and nrfA (Budiharjo 2011), supports this idea.
CLSM image of gfp-labeled knock-out mutants on Arabidopsis roots seven days after inoculation. (A) Root tip of the primary root and (B) primary root surface with a emerging lateral root colonized by FB01mut. (C) Border cells of the primary root tip, and (D) emerging lateral root colonized by the ΔpabB-mutant. (E) Border cells of primary root tip and (F) emerging lateral root colonized by ΔyusV-mutant. (G) Border cells of primary root tip and (H) emerging lateral root colonized by ΔdegU-mutant. (I) Border cells of primary root tip and (J) emerging lateral root colonized by ΔpznA-mutant. (K) Border cells of primary root tip and (L) emerging lateral root colonized by ΔnrfA-mutant. (M) Border cells of primary root tip and (N) emerging lateral root colonized by ΔRBAM017410-mutant. (O) Primary root tip with border cells colonized by ΔabrB-mutant.
Acknowledgements
The work was supported by funds of the German Ministry for Education and Research (GenoMik-Transfer, PATHCONTROL 0315654A) and the European Union’s Seventh Framework Programme (FP/2007-2013) under Grant agreement n°312117.