 |
 |
| Plant Pathol J > Volume 29(3); 2013 > Article |
Abstract
Rice stripe virus (RSV), the type member of the genus Tenuivirus, transmits by the feeding behavior of small brown planthopper (SBPH), Laodelphax striatellus. To investigate the interactions between the virus and vector insect, total RNA was extracted from RSV-viruliferous SBPH (RVLS) and non-viruliferous SBPH (NVLS) adults to construct expressed sequence tag databases for comparative transcriptome analysis. Over 30 million bases were sequenced by 454 pyrosequencing to construct 1,538 and 953 of isotigs from the mRNA of RVLS and NVLS, respectively. The gene ontology (GO) analysis demonstrated that both libraries have similar GO structures, however, the gene expression pattern analysis revealed that 17.8% and 16.8% of isotigs were up- and down-regulated significantly in the RVLS, respectively. These RSV-dependently regulated genes possibly have important roles in the physiology of SBPH, transmission of RSV, and RSV and SBPH interaction.
Rice stripe virus (RSV), the prototype virus of genus tenuivirus, is a non-enveloped RNA virus which forms circular filamentous shape of ribonucleoprotein particles with four species of segmented single-stranded RNA genome of RNA1, 2, 3 and 4 according to the size of each RNA molecule (Hibino et al., 1985; Ishikawa et al., 1989). The largest segment RNA1 encodes a 336.8 kDa protein which is thought to be a RNA dependent RNA polymerase, RdRP (Toriyama et al., 1994). RNA2 encodes 94 kDa glycoprotein and NS2 (Takahashi et al., 1993), RNA3 encodes NS3 and CP (Zhu et al., 1991), and RNA4 encodes SP and NS4 (Zhu et al., 1992). RNA1 is a negative-sense single strand while RNA2, 3, and 4 are ambisense in their coding strategy (Hamamatsu et al., 1993). RSV is one of the most serious viral rice pathogens which occur in South Korea, China, and Japan. The typical symptoms of RSV infected rice plant are chlorotic stripes or necrotic streaks on leaves, and premature wilting. Some of the leaves emerge abnormally unfolded, and then elongate to become twisted and droop. The RSV often causes significant reduction in yield. Especially the rice plants inoculated with RSV in 11-leaf stage or earlier produce no grain (Hibino, 1996; Ling, 1972).
The mechanisms of the insect vector mediated plant virus transmission are diverse based on the differences of the duration of which the vector insects can harbor infectious viral particles. The transmission mechanisms can be classified into three categories; non-persistent (harboring viruses for a few seconds/minutes), semi-persistent (a few hours/days, but lost upon molting), and persistent (often throughout the lifespan of the vectors) (Ng and Falk, 2006). Most of the plant viruses are dependent on the vector insects for plant-to-plant spread, and one of the most important vector insects are hemipterans, which can transmit viruses during feeding with their piercing-sucking mouthpart in the phloem (Hogenhout et al., 2008). The small brown planthopper (SBPH), Laodelphax striatellus Fall├®n (Hemiptera: Delphacidae), is one of the vectors of RSV and classified as a vector of persistent transmission because RSV replicates persistently in the SBPH ovary to transmit virus to the offspring transovarially (Toriyama, 1986) as well as circulatively to proliferate and escape from the midgut, salivary gland and ovary barriers for host-to-host infection (Nault and Ammar, 1989). Many of the tenuiviruses rely on planthopper vectors for transmission, and the vector specificity is high (Falk and Tsai, 1998). The SBPH is one of the most important insect vectors of RSV, however, our knowledge of the SBPH biology is limited and most of the interactions between RSV and SBPH have received less attention so far.
To investigate the RSV-SBPH interaction, we employed next generation 454-pyrosequencing technology to construct expressed sequence tag (EST) libraries from SBPH because SBPH genome sequence is not available at this moment. The pyrosequencing technology (Ronaghi et al., 1996) on the Roche 454 GS-FLX platform was used to create EST databases of RSV-viruliferous SBPH (RVLS) and non-viruliferous SBPH (NVLS), and the gene expression pattern of RVLS and NVLS were compared based on the read counts of each isotigs. We also conducted quantitative real-time PCR (qPCR) experiments with selected genes from the EST database to validate the sequencing results.
The RVLS and NVLS were collected from the RSV infected rice field and healthy rice seedlings, respectively. The RVLS and NVLS colonies were reared in the laboratory on 2 - 3 cm tall rice seedlings in glass vessels under a light:dark cycle of 16:8 h at 25┬░C and transferred to fresh seedlings every 10 - 14 days to assure sufficient nutrition. After a few generations, total RNA for nucleotide sequencing was extracted from RVLS and NVLS with TRI-Reagent (Invitrogen, USA) according to the manufacturerŌĆÖs instruction, and RT-PCR was performed to detect RSV in the RVLS and NVLS. Two pairs of primers for specific amplification of RSV sequences were designed as follows; RSV-CP-F: 5ŌĆ▓-GCT CAT AAA GCT GAT GTT G-3ŌĆ▓ and RSV-CP-R: 5ŌĆ▓-GTC TTC ACT TTC CCA TTG-3ŌĆ▓ correspond to the nucleotide sequence of coat protein gene in RNA3 with 969 base pair (bp) of expected PCR product. RSV-NCP-F: 5ŌĆ▓-GAA AGA TCC AGA GAG AGT CA-3ŌĆ▓ and RSV-NCP-R: 5ŌĆ▓-CAA AAT AAG TTG GCA TCT TC-3ŌĆ▓ correspond to the nucleotide sequence of non-capsid protein gene in RNA4 with 537 bp of expected PCR product. Another two pairs of primers for specific amplification of SBPH sequences were designed as follows; CCOS-F: 5ŌĆ▓-ATT GCT CAT AAA GCT GAT GTT G-3ŌĆ▓ and CCOS-R primer: 5ŌĆ▓-CCG TAC CAG GAC GAA TTA-3ŌĆ▓ correspond to the nucleotide sequence of cytochrome c oxidase subunit II with 684 bp of expected PCR product. ATP-F: 5ŌĆ▓-CCC TCA AGT CAC CTC TCT-3ŌĆ▓ and ATP-R: 5ŌĆ▓-ATG GAA TTT CTG AGG AA-3ŌĆ▓ correspond to the nucleotide sequence of ATP synthase F0 subunit 6 with 655 bp of expected PCR product. cDNAs were synthesized with the reverse primer of each genes, and RT-PCR reactions were performed with AccuPower one-step RT-PCR premix (Bioneer, Korea). The RT-PCR result confirmed that only the RNA samples from the RVLS amplified viral sequences while both RVLS and NVLS samples amplified SBPH specific sequences (data not shown).
To conduct the transcriptome sequencing of RVLS and NVLS, total RNA samples were extracted with TRI-Reagent from approximately 500 SBPHs of RVLS and NVLS colonies, respectively. 200 ng of mRNA samples were eluted from the total RNA samples by using oligo-dT hybridization followed by heat fragmentation, and reverse transcription to synthesize cDNA with random hexamer. Second strand was synthesized with second strand enzyme followed by 2 hour incubation in buffer for recovery of double stranded cDNA, 5ŌĆ▓-phosphorylation and addition of 3ŌĆ▓-dA overhang. Finally, adaptor was ligated and the samples were amplified to be prepared for sequencing. Approximately 5 ug of each amplified cDNA samples from RVLS and NVLS were subjected to two of 1/8 plate sequencing runs with a Roche 454-GS-FLX Titanium sequencer. The sequence data were processed with GS-FLX software v2.5.3 to remove low quality sequences, and the resulting sequences were then screened against the NCBI UniVec database and Escherichia coli genome sequences to remove contaminant sequences, then the sequences shorter than 100 bp were discarded. The processed sequences were assembled into isotigs using Newbler v 2.5.3 software to construct two EST libraries of RVLS and NVLS. Both data sets of assembled contigs and singletons from RVLS and NVLS were pooled to construct a combined sequence dataset which was supposed to have more and longer SBPH contigs. The combined dataset was used to build a new reference EST library to compare with RVLS and NVLS, and to analyze differential gene expression pattern. The features of the three EST libraries are summarized in Table 1, and the EST libraries in FASTA format are available upon request. The sequence data of RVLS and NVLS were deposited in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) with accession numbers of SRR538840 and SRR538841, respectively.
To obtain an overview of the transcriptome of SBPH, the isotigs of RVLS and NVLS were subjected to GenBank nr database with BLASTX and gene ontology (GO) analysis to annotate the isotigs into three main categories; biological process, cellular component, and molecular function (Ashburner et al., 2000). The GO distributions of the isotigs and singletons of both RVLS and NVLS were similar to each other (Table 2), and the detailed GO annotations of each subcategory also showed similar trends of distributions. This result implies that the RSV did not alter the gene expression attributes of SBPH significantly. The 2,555 isotigs of the combined dataset were analyzed by comparing with Drosophila proteins using BLASTX with a cut-off value of 1e-05 to identify their gene functions. Total of 1,467 isotigs of the BLASTX hits were subjected to Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology analysis (Kanehisa et al., 2004) to categorize their functional classes (Fig. 1A).
To compare the gene expression pattern of RVLS and NVLS, the total read counts of SBPH isotigs from RVLS and NVLS were normalized arithmetically, and then total of the 2,543 SBPH isotigs from the combined EST library were plotted based on the normalized read counts in RVLS and NVLS sequencing results. The significant differences of the transcription levels were determined by comparing the read counts of each isotig (binominal probability of < 0.1) to identify the isotigs which were up- or down-regulated by RSV. The distribution of isotigs showed that majority of the host genes were not significantly affected while 453 (17.8%) and 428 (16.8%) of isotigs were up- and down-regulated in RVLS, respectively (Fig. 2). The isotigs which showed significant differences of the transcription levels were also subjected to KEGG orthology anaylsis to show their functional classes. The isotigs related to folding, sorting and degradation, and cellular processes were up-regulated while the genes related to translation and xenobiotics metabolism were down-regulated compared with those of the NVLS. In particular, the genes related to translation were significantly down-regulated (Fig. 1B). Especially, the numbers of isotigs which showed more than ten normalized read count in either RVLS or NVLS with more than 10-fold of up-and down-regulation in RVLS were 234 and 308, respectively. These differentially expressed isotigs were subjected to GenBank insect nr database with BLASTX, and the total of 23 up-regulated and 22 down-regulated isotigs of BLASTX hits were summarized in Tables 4 and 5, respectively.
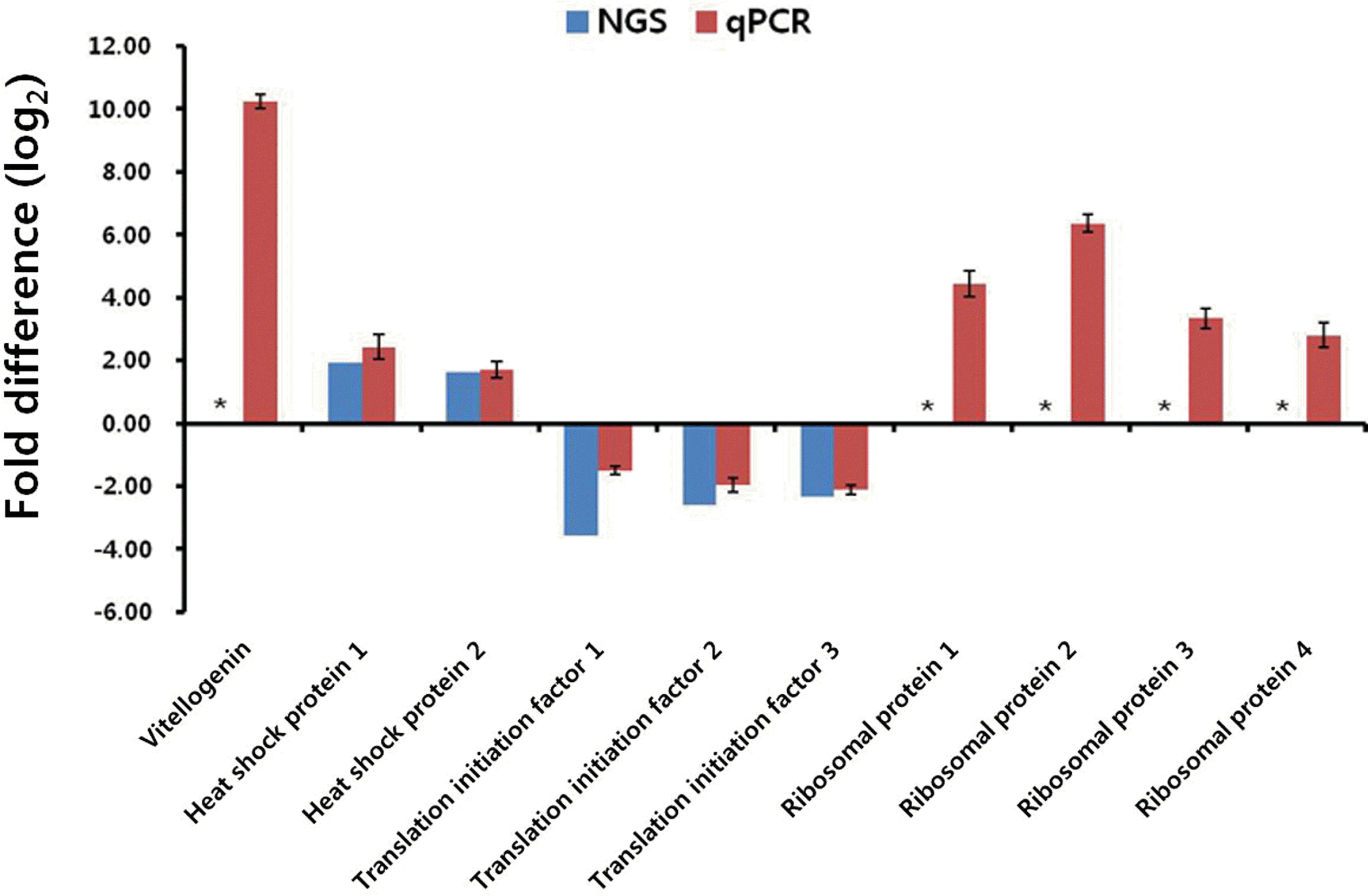
To validate the pyrosequencing results of the SBPH transcriptomes, the transcription levels measured based on the read counts were compared with those of qPCR. For qPCR, total of seven translation-related isotigs (translational initiation factor 1, 2, and 3, and ribosomal protein 1, 2, 3, and 4), which belong to the KEGG functional group of which showed the most significant difference of the transcription level between RVLS and NVLS, were selected. Also heat shock protein 1 and 2, and vitellogenin genes were selected for qPCR along with elongation factor 2 (EF2) gene as a reference for normalization (Tong et al., 2009). The cDNA was synthesized from the total RNA of RVLS and NVLS using QuantiTect Reverse Transcription Kit (QIAGEN, Germany) according to the manufacturerŌĆÖs instructions, followed by qPCR conducted using EvaGreen qPCR Mastermix (Applied Biological Materials Inc, Canada) and CFX96Ōäó Real-Time System (BIO-RAD, USA). The primers used for the qPCR are listed in Table 5. The qPCR results confirmed that the ten isotigs were indeed transcribed in both RVLS and NVLS, and the calculation of the relative transcription levels of the isotigs using 2ŌłÆ╬öCt method (Pfaffl, 2001) demonstrated that the seven genes (vitellogenin, heat shock protein 1 and 2, and ribosomal protein 1, 2, 3 and 4) and the other three genes (translation initiation factor 1, 2 and 3) were up- and down-regulated in RVLS, respectively. To calculate fold differences of the isotigs, the read counts of the isotigs in RVLS and NVLS which are the corresponding isotigs of the qPCR experiment were measured in reads per kilobase of exon per million mapped sequence reads (RPKM) (Mortazavi et al., 2008). The sequence reads of the five isotigs (heat shock protein 1 and 2, translation initiation factor 1, 2 and 3) were confirmed in both RVLS and NVLS while the other five isotigs (vitellogenin, ribosomal protein 1, 2, 3 and 4) were confirmed in RVLS exclusively. The expression levels measured by qPCR were in a good accordance with the ratios of read counts of the EST libraries (Fig. 3).
The RSV transcripts of three of the seven viral ORFs, NS3, CP and RdRP were confirmed in RVLS, and their sequence read counts were 293, 224 and 5, respectively. The total RSV read counts were 522, and this was 0.63% of the total of 83,041 valid sequence reads of RVLS contigs and singletons. The low read count of RdRP suggests that the replication of RSV is not highly active in SBPH while NS3 was the most abundant RSV transcript in RVLS which codes for viral suppressor of siRNA (Hemmes et al., 2009). CP was the second most abundant RSV transcript which codes for capsid protein (Hayano et al., 1990). Both NS3 and CP are the genes located on genomic and antigenomic strand of RSV RNA3, respectively. Therefore the similar levels of NS3 and CP transcripts possibly suggest that the genes in RNA3 are under the same gene expression regulation mechanism, however, qPCR experiments to quantify the RSV transcripts in RVLS need to be performed to prove the hypothesis.
The 454 pyrosequencing results demonstrated that the next generation sequencing (NGS) technique was able to reflect the characteristics of the transcriptome of RVLS. The genes in the RVLS which showed different transcription patterns indicated that the RSV may alter the gene expression of the SBPH which possibly affects the physiology, fitness and behavior of RVLS. The up-regulated heat shock protein genes might be one of the examples that the RSV may take the advantage of altering gene transcription to improve fitness of the vector because heat shock proteins play a significant role in generating tolerance to stresses (Krebs, 1999; Sorensen et al., 2003). It is also interesting to note that the transcription level of vitellogenin in RVLS was highly up-regulated, which means RSV might modulate the host vitellogenin pathway genes to promote the oogenesis or alter sex ratio of RVLS to facilitate its persistent transovarial transmission via the offsprings of RVLS. The highly up-regulated ribosomal protein genes were also the possible consequence of the up-regulation of vitellogenin because the ovarian cells need more amount of translational machinery, however, the down-regulated translation initiation factors are paradoxical because the recruitment of transcription initiation factors is a crucial step of protein synthesis. Many viruses can inhibit host translation initiation factors to take over host gene expression by which are usually viral protein mediated protein level interactions, rather than transcription regulation (Walsh and Mohr, 2011). It suggests that RSV possibly can alter the transcription of the genes for cellular translational machinery direct or indirect manner. Another example of viral modulation of cell physiology is the major viral transcript NS3, a RNAi suppressor which binds 21 nt siRNA, and it might be a viral strategy to evade defense system of SBPH and rice plant (Alvarado and Scholthof, 2009; Hemmes et al., 2007). Early studies on vector transmission of plant viruses demonstrated the specificity of the virus-vector interaction, which says that the instances of the biological properties associated with the transmission process may differ significantly (Ng and Falk, 2006). However, the genomics approach to study the virus-vector interaction using NGS technique might contribute to investigate the gene regulation mechanism in the persistent vector insects and the upstream regulation factors, which will be a new approach to study the interaction among the virus, plant, and vector.
Acknowledgments
This work was supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ008198), Rural Development Administration, Republic of Korea. Joo Hyun Lee and Xue Ying Tao were supported by the second stage of the Brain Korea 21 project.
Fig.┬Ā1
KEGG analysis of total (A), and UP and DOWN (B) isotigs. (A) Functional groups of the combined EST database defined by KEGG orthology were classified as carbohydrate metabolism, energy metabolism, lipid metabolism, nucleotide metabolism, amino acid metabolism, biosynthesis of other secondary metabolites, xenobiotics biodegradation and metabolism, transcription, translation, folding sorting and degradation, replication and repair, membrane transport, signal transduction, cellular processes, and organismal systems. (B) The isotigs which showed significantly different transcription level based on the read number (a binominal probability of < 0.1) were classified as UP or DOWN. UP and DOWN indicate the genes up- and down-regulated in RVLS, respectively.
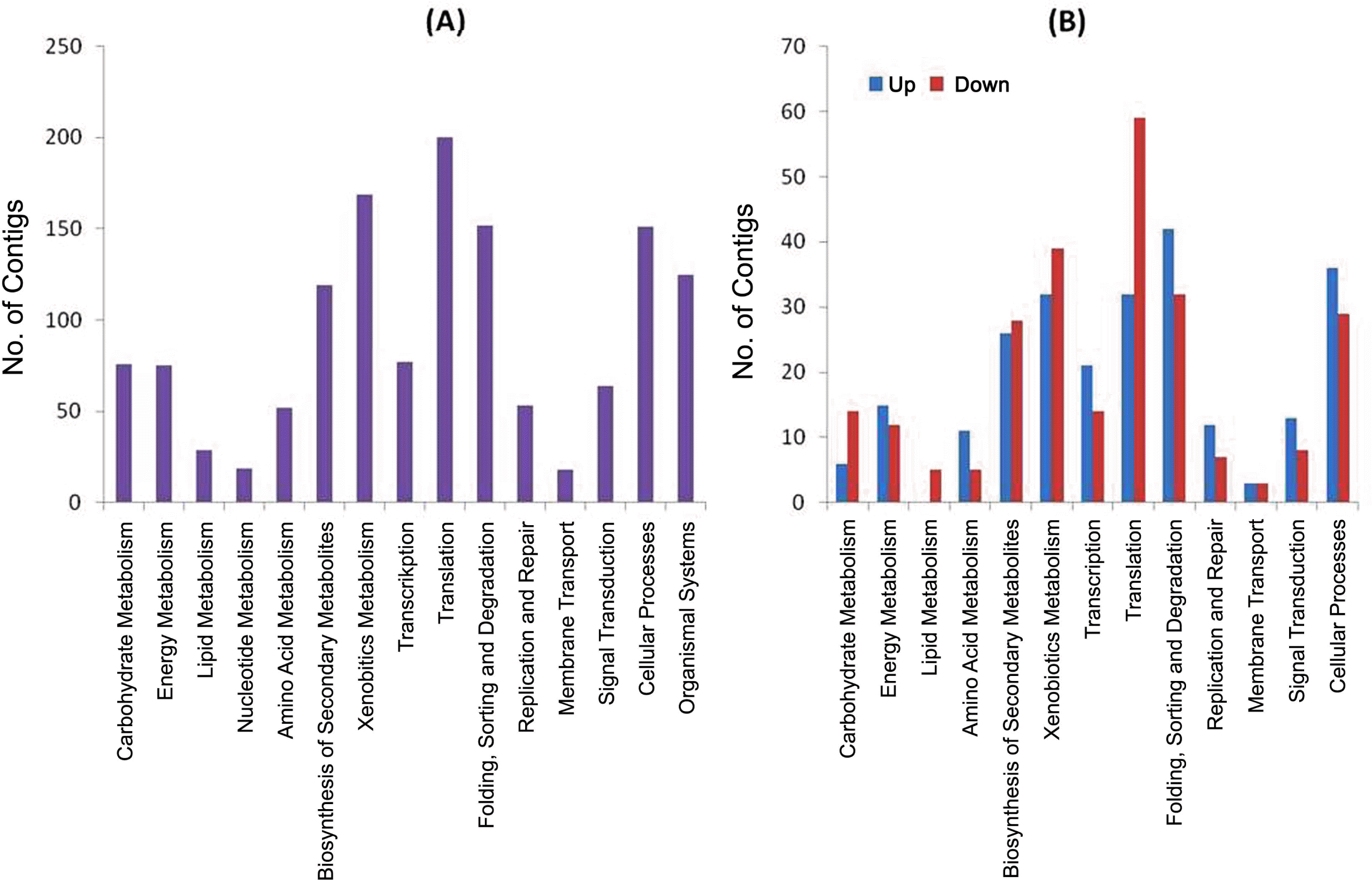
Fig.┬Ā2
Graph of the read counts of RVLS and NVLS for each isotig. The sequence read counts of RVLS and NVLS of each isotig were graphed on an x-y plot. For convenience, isotigs were ploted on two separated graphs. For the contigs which have less than 200 read counts are shown in the left panel, and the isotigs with more than 200 read counts were graphed in the write panel. Total of 2,543 SBPH isotigs were subjected to this plot, and 1,662 of the isotigs were not significantly affected (p > 0.1) while 453 and 428 of isotigs were up-and down-regulated, respectively. Up- and down-regulated isotigs are indicated as diamonds and squares, respectively.
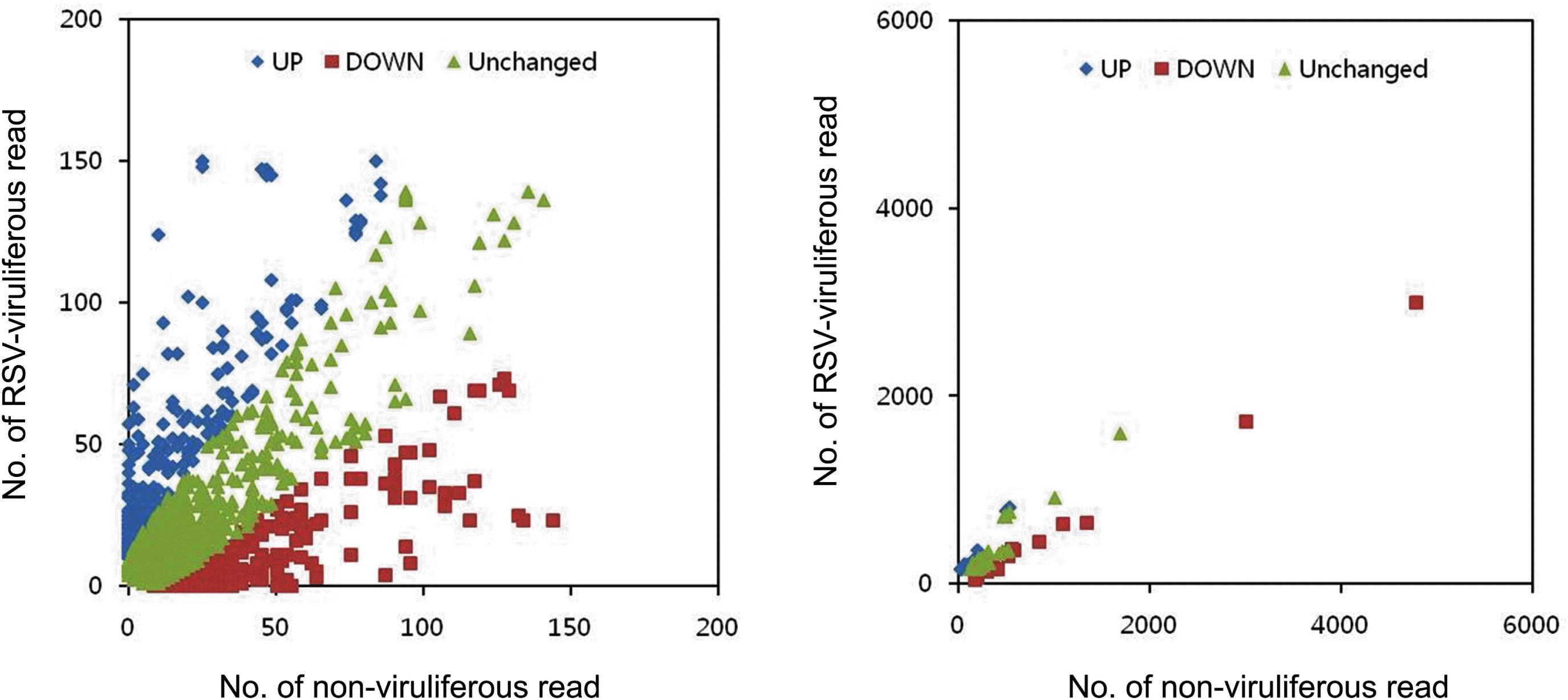
Fig.┬Ā3
Validation of the 454 pyrosequencing results by qPCR. The expression profiles of the ten isotigs were analyzed by qPCR to validate the 454 pyrosequencing results. The relative transcription levels are expressed in fold differences of RVLS to NVLS calculated by qPCR using 2ŌłÆ╬öCt method. Vitellogenin, heat shock protein 1 and 2, and ribosomal protein 1, 2, 3 and 4 were up-regulated while translation initiation factor 1, 2 and 3 were down-regulated in RVLS. The transcription level calculated with sequence read counts of the five of the ten genes, heat shock protein 1 and 2, translation initiation factor 1, 2 and 3, demonstrated that the transcription levels measured by genomics and experimental methods were in a good agreement. The other five genes, vitellogenin, ribosomal protein 1, 2, 3 and 4, which showed substantial read counts in RVLS, had no read counts in NVLS so that these genes were not able to calculate the fold differences (indicated with asterisk). NGS and qPCR indicate the fold differences calculated with the sequencing and qPCR results, respectively.
Table┬Ā1
Summary of the features of Laodelphax striatellus EST libraries
Table┬Ā2
BlastX search and Gene Ontology analysis of Laodelphax striatellus EST libraries
Table┬Ā3
The genes up-regulated more than 10-fold in RSV viruliferous Laodelphax striatellus
Table┬Ā4
The genes down-regulated more than 10-fold in RSV viruliferous L. striatellus
Table┬Ā5
Primers used for quantitative real-time PCR expression profiling of putative transcripts generated from 454-pyrosequencing
References
Alvarado, V and Scholthof, HB 2009. Plant responses against invasive nucleic acids: RNA silencing and its suppression by plant viral pathogens. Semin. Cell Dev. Biol. 20:1032-1040.



Ashburner, M, Ball, CA, Blake, JA, Botstein, D, Butler, H, Cherry, JM, Davis, AP, Dolinski, K, Dwight, SS, Eppig, JT, Harris, MA, Hill, DP, Issel-Tarver, L, Kasarskis, A, Lewis, S, Matese, JC, Richardson, JE, Ringwald, M, Rubin, GM, Sherlock, G and Consortium, GO 2000. Gene ontology: Tool for the unification of biology. Nat. Genet. 25:25-29.




Falk, BW and Tsai, JH 1998. Biology and molecular biology of viruses in the genus tenuivirus. Annu. Rev. Phytopathol. 36:139-163.


Hamamatsu, C, Toriyama, S, Toyoda, T and Ishihama, A 1993. Ambisense coding strategy of the Rice stripe virus genome: In vitro translation studies. J. Gen. Virol. 74(Pt 6):1125-1131.


Hayano, Y, Kakutani, T, Hayashi, T and Minobe, Y 1990. Coding strategy of Rice stripe virus: Major nonstructural protein is encoded in viral RNA segment 4 and coat protein in RNA complementary to segment 3. Virology. 177:372-374.


Hemmes, H, Kaaij, L, Lohuis, D, Prins, M, Goldbach, R and Schnettler, E 2009. Binding of small interfering RNA molecules is crucial for RNA interference suppressor activity of Rice hoja blanca virus ns3 in plants. J. Gen. Virol. 90:1762-1766.


Hemmes, H, Lakatos, L, Goldbach, R, Burgyan, J and Prins, M 2007. The ns3 protein of Rice hoja blanca tenuivirus suppresses RNA silencing in plant and insect hosts by efficiently binding both sirnas and mirnas. RNA. 13:1079-1089.



Hibino, H, Usugi, T, Omura, T, Tsuchizaki, T, Shohara, K and Iwasaki, M 1985. Rice grassy stunt virus - a planthopper-borne circular filament. Phytopathology. 75:894-899.

Hogenhout, SA, Ammarel, D, Whitfield, AE and Redinbaugh, MG 2008. Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 46:327-359.


Ishikawa, K, Omura, T and Hibino, H 1989. Morphological-characteristics of Rice stripe virus. J. Gen. Virol. 70:3465-3468.

Kanehisa, M, Goto, S, Kawashima, S, Okuno, Y and Hattori, M 2004. The kegg resource for deciphering the genome. Nucleic Acids Res. 32:D277-D280.



Krebs, RA 1999. A comparison of hsp70 expression and thermotolerance in adults and larvae of three drosophila species. Cell Stress Chaperon. 4:243-249.

Ling, KC 1972. Rice virus diseases. Los Banos, Philippines. International rice research institute.
Mortazavi, A, Williams, BA, Mccue, K, Schaeffer, L and Wold, B 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods. 5:621-628.



Nault, LR and Ammar, E 1989. Leafhopper and planthopper transmission of plant-viruses. Annu. Rev. Entomol. 34:503-529.

Ng, JC and Falk, BW 2006. Virus-vector interactions mediating nonpersistent and semipersistent transmission of plant viruses. Annu. Rev. Phytopathol. 44:183-212.


Pfaffl, MW 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45.



Ronaghi, M, Karamohamed, S, Pettersson, B, Uhlen, M and Nyren, P 1996. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 242:84-89.


Sorensen, JG, Kristensen, TN and Loeschcke, V 2003. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 6:1025-1037.

Takahashi, M, Toriyama, S, Hamamatsu, C and Ishihama, A 1993. Nucleotide sequence and possible ambisense coding strategy of Rice stripe virus RNA segment 2. J. Gen. Virol. 74:769-773.


Tong, ZG, Gao, ZH, Wang, F, Zhou, J and Zhang, Z 2009. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC. Mol. Biol. 10:71.




Toriyama, S 1986. Rice stripe virus: Prototype of a new group of viruses that replicate in plants and insects. Microbiol. Sci. 3:347-351.

Toriyama, S, Takahashi, M, Sano, Y, Shimizu, T and Ishihama, A 1994. Nucleotide sequence of RNA 1, the largest genomic segment of Rice stripe virus, the prototype of the tenuiviruses. J. Gen. Virol. 75:3569-3579.


Walsh, D and Mohr, I 2011. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 9:860-875.




- TOOLS
-
METRICS

-
- 16 Crossref
- 0 Scopus
- 6,048 View
- 15 Download
- Related articles
-
Transcriptome Analysis of the Barley
-Rhynchosporium secalis Interaction2014 December;30(4)



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print



