The RpoS Sigma Factor Negatively Regulates Production of IAA and Siderophore in a Biocontrol Rhizobacterium, Pseudomonas chlororaphis O6
Article information
Abstract
The stationary-phase sigma factor, RpoS, influences the expression of factors important in survival of Pseudomonas chlororaphis O6 in the rhizosphere. A partial proteomic profile of a rpoS mutant in P. chlororaphis O6 was conducted to identify proteins under RpoS regulation. Five of 14 differentially regulated proteins had unknown roles. Changes in levels of proteins in P. chlororaphis O6 rpoS mutant were associated with iron metabolism, and protection against oxidative stress. The P. chlororaphis O6 rpoS mutant showed increased production of a pyoverdine-like siderophore, indole acetic acid, and altered isozyme patterns for peroxidase, catalase and superoxide dismutase. Consequently, sensitivity to hydrogen peroxide exposure increased in the P. chlororaphis O6 rpoS mutant, compared with the wild type. Taken together, RpoS exerted regulatory control over factors important for the habitat of P. chlororaphis O6 in soil and on root surfaces. The properties of several of the proteins in the RpoS regulon are currently unknown.
Some rhizobacterial strains have been widely used as biocontrol agents, because they possess beneficial traits, such as the production of plant growth hormones and other secondary metabolites (Kim et al., 2011). An aggressive rhizobacterium, Pseudomonas chlororaphis O6 produces secondary metabolites, including phenazines and pyrrolintrin, which directly inhibit the growth of plant pathogenic fungi (Kang et al., 2007; Park et al., 2011). P. chlororaphis O6 also produces an iron-chelating pyoverdine-like (PVD) siderophore (Dimka et al., 2011) and the plant growth hormone, indole acetic acid (IAA) (Dimka et al., 2012; Kang et al., 2004), which affects plant growth (Beyeler et al., 1999; Costacurta and Vanderleyden, 1995; Patten and Glick, 2002; Vansuyt et al., 2007). Root colonization of P. chlororaphis O6 induces systemic resistance against viral, bacterial, and fungal plant diseases, as well as drought stress (Cho et al., 2008; Han et al., 2006; Kim et al., 2004; Ryu et al., 2007; Spencer et al., 2003).
In plant-associated pseudomonads, the sigma factor RpoS controls the expression of genes promoting survival under stress conditions (Kang et al., 2004; Miller et al., 2001). In P. chlororaphis O6, the rpoS transcript was influenced by a ppk gene encoding a polyphosphate kinase (Kim et al., 2007). The rpoS gene in P. putida is required for production of oxidative stress related enzymes, such as manganese superoxide dismutase and catalases, and for root colonization (Miller et al., 2001). RpoS also plays an important role in the survival of P. fluorescens Pf-5 on plant surfaces (Sarniguet et al., 1995). We used a proteomic approach to initiate a better understanding of the role of RpoS as a regulator in P. chlororaphis O6. The previously constructed rpoS mutant and its complemented rpoS mutant (Oh et al., 2012), as well as a gacS and complemented gacS mutant (Spencer et al., 2003), were used in this study. Previous studies showed the Gac system in P. chlororaphis O6 to regulate expression from rpoS (Kang et al., 2004). Strains were preserved in King’s B medium (KB) or Luria Bertani (LB) broth containing 15% glycerol (v/v) at −80 °C.
Cell extracts were prepared by sonication of cells grown to stationary phase cells in LB and centrifugation at 15,000 × g for 10 min. Protein concentration was measured using the BioRad Protein Assay kit (BioRad, Hercules, CA, USA). The proteins were separated by isoelectric focusing (IEF) at 20 °C with a pH gradient (4 – 10) using a Multiphor II electrophoresis unit (Amersham Biosciences, Piscataway, NJ, USA). Sodium dodecyl sulfate-PAGE with 10 – 16% gradient gel was performed using a Hoefer DALT 2D system (Amersham Biosciences, Piscataway, NJ, USA) and the gels were silver-stained as described by Oakley et al. (1980) with the omission of glutaraldehyde. Quantitative analysis of the digitized images was carried out using the PDQuest (BioRad) software. Three independent 2-D gel analyses were performed, and spots with p-values of < 0.05 were selected for Q-TOF analysis.
The protein spots were digested enzymatically in-gel with 0.2 μg of modified trypsin (Promega, Madison, WI, USA) for 45 min on ice (Shevchenko et al., 1996) and were desalted on columns, 100 – 300 nL of porous reverse-phase R2 material (20 – 30 μm bead size, PerSeptive Biosystems, MA). The peptides were eluted with 1.5 μL of 50% methanol/49% H2O/1% formic acid into a pre-coated borosilicate nano-electrospray needle (Micromass, Manchester, UK) and the MS/MS data obtained with a nano-ESI on a Q-TOF2 mass spectrometer (Micromass, Manchester, UK). The MS/MS spectra were searched against the protein sequences from the NCBI databases using the MASCOT search program (www.matrixscience.com).
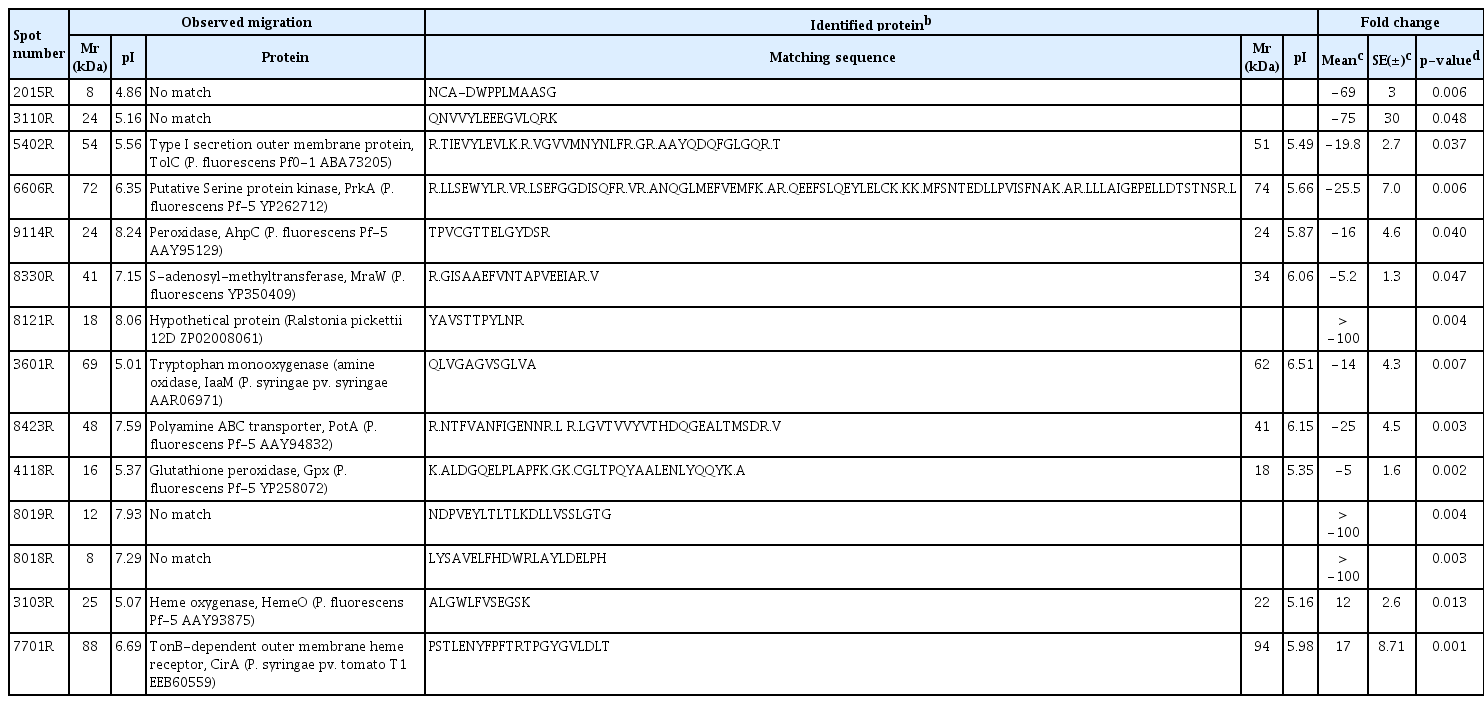
More than 700 protein spots were reproducibly detected in each gel, and representative images from P. chlororaphis O6 wild type and rpoS mutant are shown in Fig. 1. From the gels, we identified 14 spots that had a more than two fold change (P < 0.05) in levels in the rpoS mutant compared to the wild type (Fig. 1). Of those peptides, no matches were detected for four proteins (Table 1) and another corresponded to a hypothetical protein found in a Ralstonia isolate. A tryptophan monooxygenase (IaaM), outer membrane protein (TolC), which is potentially involved in export, other proteins involved in oxidative stress [peroxidase (AphC) and glutathione peroxidase (Gpx)], and at growth [polyamine transporter (PotA)], were signiticantly decreased in the rpoS mutant. Two proteins with the potential to function in cell signaling, S-adenoysyl methyltransferase (MraW) and a putative serine protein kinase (PrkA), were down-regulated in the rpoS mutant. In contrast, two proteins involved in the cellular utilization of iron from heme sources, heme oxygenase (HemeO) and a TonB-dependent membrane heme receptor (CirA) were detected at higher levels in the mutant (Table 1). RpoS regulated genes and proteins were identified in Geobacter sulfurreducens and Burkholderia pseudomallei (Nunez et al., 2006; Osiriphun et al., 2009). As expected, most of the RpoS regulated genes and proteins are related to stress responses, but many of the RpoS regulated genes and proteins are involved in the expression of diverse physiological traits, such as general metabolism, transcription, translation, cell motility and secretions (Nunez et al., 2006; Osiriphun et al., 2009).
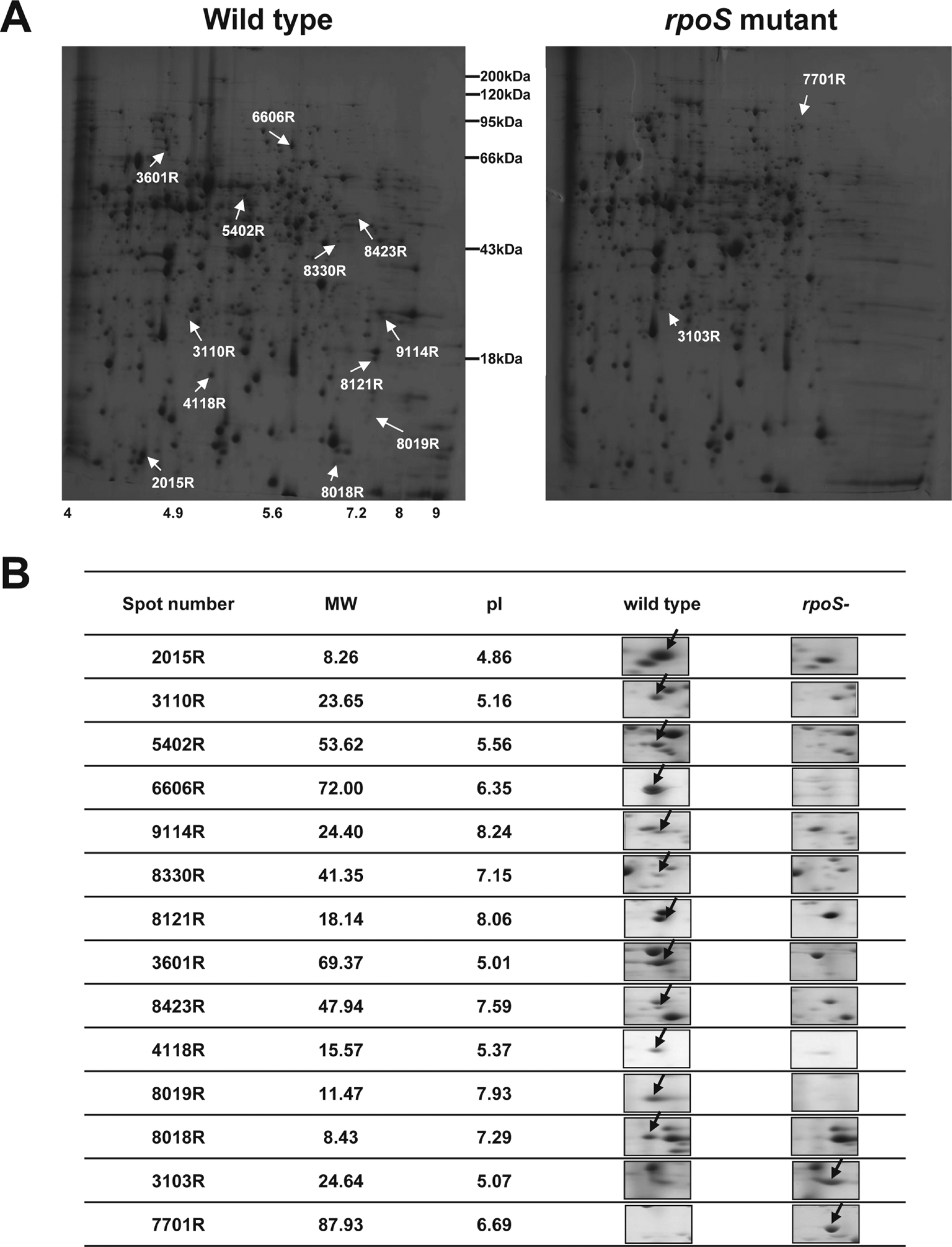
Comparison of intensities of proteins spots on two-dimensional gel electrophoresis (2-DE), in which total proteins isolated from stationary cells of P. chlororaphis O6 and rpoS mutant were loaded. (A) Representative 2-DE maps of P. chlororaphis O6 wild type and rpoS mutant. Total proteins of wild type and rpoS mutant were separated by gradient isoelectric focusing with a pH and SDS-PAGE, and then the gels were stained with silver. (B) The magnified protein spots showed differential expression between P. chlororaphis O6 and rpoS mutant. Arrows indicate differentially expressed proteins between wild type and rpoS mutant in the three independent 2-DE analyses.
To understand the significance of these changes, the production of the plant hormone IAA was compared between the rpoS mutant and the wild type strains. Cells were grown for 48 h in the low-iron medium described by Dimkpa et al. (2012) amended with 200 μM tryptophan for the IAA studies. IAA was detected in cell free culture filtrates by the Salkowski colorimetric assay (Patten and Glick, 2002), using the authentic compound as the standard and was authenticated by HPLC as described in Dimkpa et al. (2012). Production of IAA was increased in the rpoS mutant. The highest level was similar to those observed from cultures of the gacS mutant (Fig. 2A) suggesting that RpoS was the major regulator. Although P. chlororaphis O6 is reported to produce IAA in the presence of tryptophan (Kang et al., 2006), the pathway was not elucidated clearly. A recent study indicated that the indole-3-acetamide pathway is involved for IAA production in P. chlororaphis O6 (Dimkpa et al., 2012). In this study, it was unexpected that transcripts from iaaM, encoding the first enzyme, tryptophan monooxygenase, potentially involved in conversion of tryptophan to indole-3-acetamide was down regulated in the rpoS mutant (Table 1). We are currently constructing the iaaM defective mutant and transcriptional analysis of the iaaM transcript in wild type and rpoS mutant to resolve the uncertainty between the proteomic result and phenotype in IAA production.
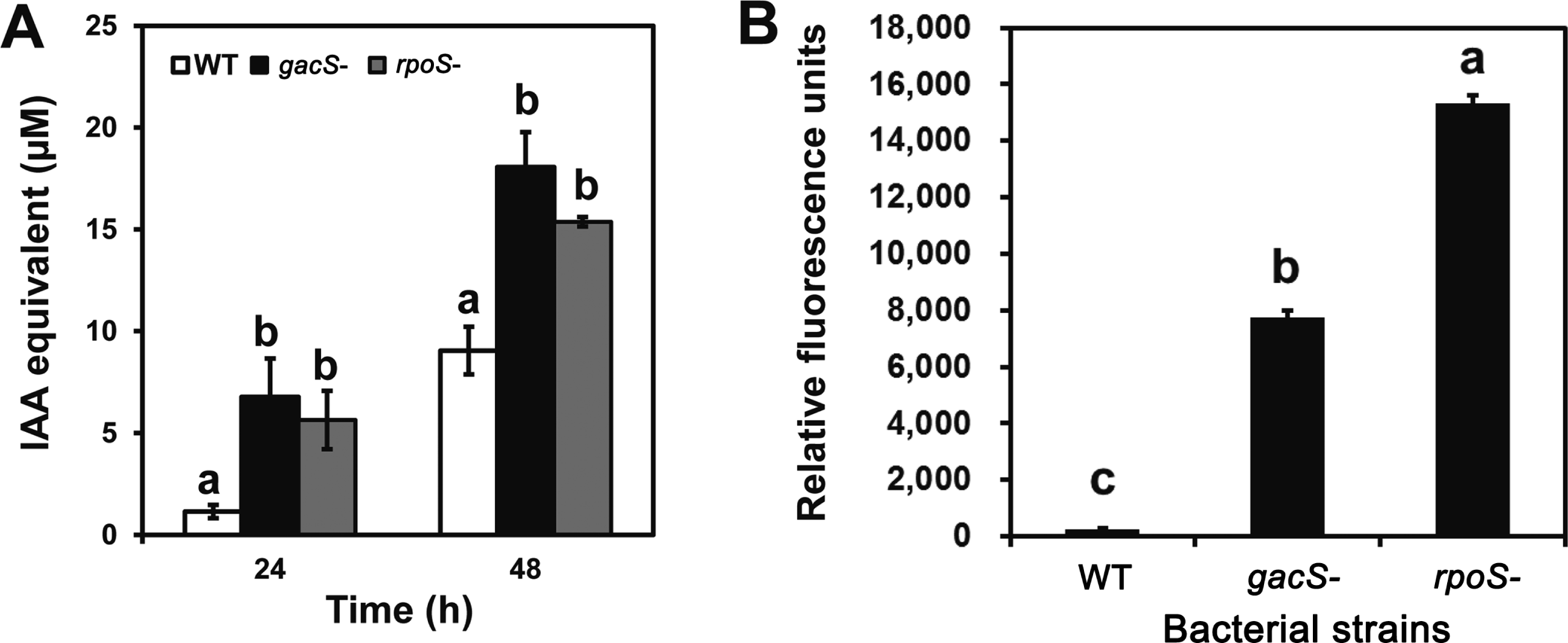
Mutations in rpoS and gacS in P. chlororaphis O6 increased IAA production (A) and PVD-like fluorescent siderophore production (B) over that of the wild type strain (WT). (A) IAA was detected in supernatants generated after growth for 24 h, and 48 h: authentic IAA was used as the standard in the Salkowski assay. (B) Siderophore levels were assessed by the characteristic fluorescence of the PVD-like siderophore with exelement-citation at 398 nm and emission at 460 nm. The data for the relative fluorescent units are means from two studies, each with two replicates; standard deviation is shown. Different letters represent significant differences at each time point (P < 0.05) based on at least two separate studies.
To understand altered iron metabolism, the production of the fluorescent siderophore was compared between the rpoS mutant and the wild type strains. Cells were grown for 48 h in low-iron medium (Dimkpa et al., 2011) with and without 100 μM Fe3+. Siderophore levels were assessed in the cell free culture by exelement-citation at 398 nm and measuring fluorescence emission at 460 nm (Dimkpa et al., 2011). Production of the PVD-like siderophore was greatly elevated in the rpoS mutant compared to that in wild type strain, with the increased production of the rpoS mutant being significantly greater than that of the gacS mutant (Fig. 2B). Growth of the strains in medium amended with 100 μM Fe3+ eliminated the characteristic PVD fluorescence, as anticipated from iron-repression of siderophore production.
Proteins in cells extracts were separated by non-denaturing electrophoresis and were stained for activities of peroxidase, catalase, and superoxide distmutase as described previously (Katsuwon and Anderson, 1990; MacAdam et al., 1992). Activity staining of extracts from late-and stationary-phase cells revealed isozymes with both catalase and peroxidase activities (Fig. 3A). Extracts from the rpoS and gacS mutants, lacked the same isozymes, whereas these activities were restored in the complemented mutants. The hydrogen peroxide-sensitive iron superoxide dismutase isozyme was produced constitutively by each strain (Fig. 3A). However, the rpoS and gacS mutants over-expressed a manganese superoxide dismutase isozyme, which was not inhibited by hydrogen peroxide. Expression of the Mnisozyme was suppressed in the complemented strains (Fig. 3A).

Effect of mutation in rpoS and gacS genes in P. chlororaphis O6 on catalase, peroxidase, and superoxide dismutase (SOD) isozyme patterns (A) and hydrogen peroxide sensitivity (B). Enzyme activities were detected after non-denaturing polyacrylamide gel electrophoresis of 100 μg of protein from extracts of cells grown in KB broth to the late log-phase (OD600nm = 1.8), or stationary-phase (OD600nm = 2.4). The results are typical of three independent experiments. (B) Hydrogen peroxide (4.5 mM final concentration) was added to P. chlororaphis O6 strains grown to the late log-phase (OD600nm = 1.8) in liquid KB cultures. Serial dilutions were performed with the culture and were plated to determine cell culturability at defined times. Error bars represent the standard deviations of three independent experiments. ComrpoS−, rpoS− mutant complemented with rpoS gene.
To measure hydrogen peroxide sensitivity, cells were grown in liquid KB medium at 28 °C to late-log phase (OD600nm= 1.8) when 4.5 mM hydrogen peroxide was added. Serial dilutions of the culture were made and were plated onto LB agar plates containing kanamycin (50 μg/mL) for the rpoS mutant and kanamycin and gentamycin (12.5 μg/mL) for the complemented mutant at the defined times. Colonies were counted after 2 days of incubation at 28 °C. Three independent experiments were evaluated for each strain. Recovery of cells exposed to hydrogen peroxide was lowest for the rpoS mutant and was restored to the wild type level by rpoS complementation (Fig. 3B).
These proteomic and metabolic studies demonstrated that RpoS regulated several proteins in P. chlororaphis O6. The proteins under RpoS control included specific isozymes of catalase, peroxidases and superoxide dismutase involved in protection against oxidative stress. The greater sensitivity to hydrogen peroxide displayed by the rpoS mutant showed the functionality of these and other changes, such as the polyamine transporter PotA (Jung and Kim, 2003; Tkacheko et al., 2001), the AhpC peroxidase (Hishinuma et al., 2006), and the Gpx glutathione peroxidase (Lan et al., 2010). These findings of altered levels of proteins involved in oxidative stress and increased sensitivity agree with observations of rpoS mutants of P. aeruginosa and P. fluorescens (Heeb et al., 2005; Suh et al., 1999). The role of RpoS in generating resistance to oxidative stress could be important in counteracting the oxidative stress imposed as the bacterium interacts with the root surface (Kang et al., 2004; Katsuwon and Anderson, 1990; Kim et al., 2000, 2004; Miller et al., 2001).
Maintenance of cellular Fe levels is essential in the control of oxidative stress. Effects of the rpoS mutation on iron homeostasis is probable because the proteomic analysis revealed accumulations of a heme-uptake receptor and a heme oxygenase and the mutant displayed enhanced secretion of a Fe(III)-binding fluorescent siderophore. Earlier, Suh et al. (1999) also found that an rpoS mutant in P. aeruginosa had increased siderophore production.
RpoS regulation of other factors involved in rhizosphere colonization is extended to IAA synthesis; IAA production was increased in the rpoS mutant. This finding agreed with our previous findings that production of IAA in P. chlororaphis O6 was negatively regulated by a two component sensor kinase, GacS (Kang et al., 2004; 2006). However, this finding did not correlate with the observation that transcript accumulation from iaaM, encoding the monooxygenase involved in transformation of tryptophan to IAA, was down-regulated according to the proteomic finding. One possibility is that the rpoS mutant prematurely down regulated iaaM transcription compared with the wild type where stationary phase repression has been noted (Dimkpa et al., 2012). Another possibility is that there is an alternate mechanism for IAA synthesis. Three bacterial pathways for IAA production from tryptophan are reported, namely the indole-3-pyruvate (IPyA), the tryptophan-side chain oxidase pathway, and the IaaM pathway (Beyeler et al., 1999; Costacurta and Vanderleyden, 1995; Patten and Glick, 2002).
Our observations showed that whereas RpoS was the major regulator for IAA formation in P. chlororaphis O6, both RpoS and the Gac sytem regulated siderophore production. Regulatory control by both RpoS and GacS for IAA production was found for Enterobacter cloacae (Saleh and Glick, 2001). Likewise, RpoS was a key regulator for IAA and siderophore production in the endophytic plant growth-stimulating Burkholderia phytofirmans (Sun et al., 2009).
These findings illustrate the pivotal role of RpoS in the regulation of factors involved in root colonization and interaction for P. chlororaphis O6. Like other microbes the RpoS-regulated proteins include those involved in oxidative stress and iron homeostasis. However, other as yet uncharacterized protein products were also RpoS-regulated and their function is currently under analysis.
Acknowledgements
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011-0011555) and the Agricultural Experiment Station at Utah State University.