Occurrence of dsRNA Mycovirus (LeV-FMRI0339) in the Edible Mushroom Lentinula edodes and Meiotic Stability of LeV-FMRI0339 among Monokaryotic Progeny
Article information
Abstract
dsRNA was found in malformed cultures of Lentinula edodes strain FMRI0339, one of the three most popular sawdust cultivated commercial strains of shiitake, and was also found in healthy-looking fruiting bodies and actively growing mycelia. Cloning of the partial genome of the dsRNA revealed the presence of the RdRp sequence of a novel L. edodes mycovirus (LeV), and sequence comparison of the cloned amplicon showed identical sequences sequence to known RNA-dependent RNA polymerase genes of LeV found in strain HKA. The meiotic stability of dsRNA was examined by measuring the ratio of the presence of dsRNA among sexual monokaryotic progeny. More than 40% of the monokaryotic progeny still contained the dsRNA, indicating the persistence of dsRNA during sexual reproduction. Comparing the mycelia growth of monokaryotic progeny suggested that there appeared to be a tendency toward a lower frequency of virus incidence in actively growing progeny.
Since the first definitive description of mycovirus more than 50 years ago (Hollings, 1962), the presence of viruses in fungi becomes rather general than exceptional (Dawe and Nuss, 2001; Ghabrial, 1998; Pearson et al., 2009). The majority of characterized mycoviruses have double-stranded RNA (dsRNA) genomes, although single-stranded RNA (ssRNA) mycoviruses are still being discovered at a significant rate. As a rule, mycovirus infections do not cause measurable phenotypic changes in the fungal hosts, and if any, remain cryptic or latent. In some cases, however, viral infection can result in considerable morphological and physiological changes, including debilitation- and virulence-related phenotypes (Castro et al., 2003; Dawe and Nuss, 2001).
Lentinula edodes (Berk.) Pegler, the shiitake, is the second most popular edible mushroom on the world market (Hadeler, 1995). Shiitake contains polysaccharides and substances for immune enhancement and antitumor activity (Hobbs, 2003). In addition, due to its ability to degrade the lignin component of hard-wood, its potential application as a biorefinery and bioremediation agent has been studied (Couto and Herrera, 2006; Okeke et al., 1993).
L. edodes is widely cultivated and managed throughout Korea. Consequently, L. edodes is one of the two most important commercial mushrooms in terms of yield and economic value (Ko et al., 2012). Traditionally, in Korea, cultivation of L. edodes was carried out by inoculating mycelia spawn on oak logs. This labor-intensive method, however, has gradually been replaced by indoor cultivation using bag cultures with a sawdust-based medium (Ko et al., 2012). Currently in Korea, using sawdust bag culture, the mycelia are fully grown in the substrate for approximately 2 months; then, another 1 or 1.5 months are required until browning and stiffening of the medium surface, which is an important marker for the development of the normal fruiting body during the following stage of cultivation. Therefore, compared to other cultivated mushrooms, such as Flammulina velutipes and Pleurotus ostreatus, the cultivation time for L. edodes is relatively long, and the culturing technique is more critical than for others. As the indoor cultivation of shiitake mushrooms continues to increase in Korea, various abnormal symptoms, such as imperfect browning of the medium surface, fluffy mycelia growth on the surface, and malformations of fruiting bodies, have been observed during cultivation and these abnormalities have resulted in serious economic losses. Therefore, along with other cultural practices, breeding of this fungus for the indoor cultivation has gained more attentions. Breeding of L. edodes has been carried out especially in Japan (Hasebe, 1991) to result in more than one hundred cultivars that have been registered to plant variety protection system in Japan. Recently, several progresses on the development of new cultivars of this mushroom have been achieved in Korea and China (Liu et al., 2010; Ryu et al., 2009).
There were several early reports of three morphologically distinct virus infections in L.edodes (Inouye et al., 1970; Mori and Mori, 1974; Ushiyama, 1983; Yamashita et al., 1975). Rytter et al. (Rytter et al., 1991) reported the presence of dsRNA in 23 out of 25 genetically-diverse lines in the USA, and a total of 14 unique electrophoretic dsRNA patterns were found in these 23 lines, which suggested widespread viral infection in shiitake mushrooms. In addition, the molecular characterization of a new linear uncapsidated dsRNA virus (LeVHKB) from an asymptomatic isolate in Japan was also reported (Magae, 2012). During the preparation of our manuscript, L. edodes spherical virus (LeSV), which showed a highly similar but different RdRp sequence from both L. edodes strains HKA and HKB, has been reported (Won et al., 2013). High incidence (66.7%) of LeSV infection and stable meiotic inheritance (90%) of LeSV were also observed. However, the relationships of these mycoviruses with any harmful effects on shiitake mushroom have not been determined because these mycoviruses are commonly found in healthy and asymptomatic fruiting bodies.
Browning is a typical symptom observed in mushroom virus X (MVX) disease in Agaricus bisporus (Rao et al., 2007), and the association of brown discoloration in the winter mushroom Flammulina velutipes with the presence of F. velutipes browning virus (FvBV) was also reported (Magae and Sunagawa, 2010). In this study, during an initial survey of the causes of abnormal browning of the medium surface and fruiting body of a popular commercial line of L. edodes, we were able to confirm the presence of dsRNA. In order to understand the biological function of this dsRNA and to maintain virus-tested parental strains for breeding, we characterized this dsRNA and examined the stability of the dsRNA.
Abnormal-looking fruiting bodies of L. edodes strains were collected and provided by the Forest Mushroom Research Institute (FMRI), Yeoju, Korea. All fungal cultures were maintained on potato dextrose agar (PDA) in darkness at 25 °C with periodic transfer and a miniprep method for purifying dsRNA (Park et al., 2008) was applied to test the presence of a dsRNA. In three representative mycelia cultures of the abnormally browning, which showed limited area of browning or whole area of browning but with different density, sawdust bags of collected samples, the presence of a dsRNA fragment of around 10.1-kb was reproducibly observed. No other co-migrating dsRNA fragments were observed in a repeat trial. The presence of dsRNA was further verified by digestions using RNase A, RNase III, and DNase as described previously (Park et al., 2008). The purified dsRNA proved resistant to DNase and RNase A at a high-salt concentration (0.375 M NaCl), but disappeared upon exposure to dsRNA-specific RNase III and RNase A under low-salt concentrations (data not shown). In addition, dsRNA was also detected when either the stipes or pilus were used as sources of tissue.
In order to acquire cDNA clones that correspond to a part of the RNA-dependent RNA polymerase (RdRp) of the mycoviurs in L. edodes FMRI0339, first-strand cDNA synthesized with a random primer was used as a template for PCR amplifications with a degenerated primer pair that was designed for consensus nucleotide sequences corresponding to conserved amino acids in the specific RT-like super family domain of the mycovirus. The primers used were MV-RDRP (forward), 5′-YTD TAY WSN GGD TGG MGD–3′, and MV-RDRP (reverse), 5′-HGG HTC HCC HAR HCC HCC-3′. Using degenerate primers derived from the conserved motifs of the known fungal RdRp, a 561-bp DNA fragment was amplified, cloned into the pGEM-T Easy vector (Promega, Madison, USA), and sequenced using universal primers. Sequence analysis of the clone revealed that it showed the 100% identity to the recently deposited RdRp of the L. edodes strain HKA mycovirus (GenBank No. AB646992). It also showed high similarity, with 95% and 97% identity at the nucleotide level to the RdRp of the L. edodes HKB (GenBank No. AB429556) and L. edodes LeSV (GenBank No. JQ687140) mycoviruses, respectively, whose molecular characterization have been recently performed (Fig. 1).

Alignment of the deduced amino acid sequences of the partial RNA-dependant RNA polymerase (RDRP) proteins of Korean Lentinula edodes FMRI0339 mycovirus (LeV-FMRI0339) with other mycoviruses. The amino acid sequences of the RDRP proteins from Lentinula edodes HKA mycovirus (LeV-HKA) (GenBank Accession No. AB646992), L. edodes HKB mycovirus (LeV-HKB) (GenBank Accession No. AB429556), and L. edodes spherical virus (LeSV) (GenBank Accession No. JQ687140) were manually adjusted and shaded for the degree of conservation using Jalview. Arrows indicate amino acid residues used to design primers and primer directions.
Since L. edodes FMRI0339 is a very popular sawdust cultivated shiitake strain, we examined five healthy-looking fruiting bodies of L. edodes FMRI0339 collected from local markets, and three mycelia of L. edodes FMRI0339 from the stock center of the forest mushroom research institute (FMRI). Among those tested, all L. edodes FMRI0339 strains showed the presence of dsRNA. However, two other popular sawdust cultivated strains, such as L. edodes FMRI0985 and Chamaram, did not show the presence of dsRNA. The presence of dsRNA in L. edodes FMRI0339 was so common that we tested the meiotic stability of the dsRNA by measuring the presence of the dsRNA among monokaryotic strains originating from the basidiospore of the fruiting body of L. edodes FMRI0339. Monokaryotic strains were generated by germinating the collected basidiospores on PDA at 25 °C for 7 days. The monokaryotic stage was identified by observing the absence of the clamp connection in the mycelia of monokaryotic strains under a light microscope. As shown in Fig. 2, more than 40% (17/40) of the monokaryotic progeny contained the dsRNA. Considering the general concept that sexual reproduction in the fungal host functioned to remove mycovirus (Carbone et al., 2004; Ikeda et al., 2004), the frequency of virus incidence in sexually reproduced monokaryotic progeny was remarkably high, which suggested the presence of either different modes of viral transmission or mating mechanism in this fungus. Furthermore, considering the stable persistence of dsRNA in L. edodes FMRI0339, the possibility of underestimating the natural ability of the mycovirus to migrate within the population (Brusini and Robin, 2012), and transmission between somatically incompatible hosts under natural conditions (Vainio et al., 2013), the frequency of virus incidence has gained much attention.

(A) Ethidium bromide-stained gel showing dsRNA. Lanes a and b contain dsRNA preparations from two representative isolates of Cryphonectria parasitica UEP1 strain (CHV1) and C. nitschkei (CnV1-BS122), which contain the characteristic 12.7-kb viral genome of hypovirus CHV1-EP713 and segments of chrysovirus CnV1-BS122, respectively. Lanes 1–40 contain dsRNA preparations from corresponding monokaryotic progeny, M1-M40, of L. edodes FMRI0339. (B) Northern blot analysis of dsRNA using the cloned 561-bp fragment representing a part of the RdRp of the L. edodes HKA mycovirus. Arrows indicate the mycovirus from monokaryotic progeny of L. edodes FMRI0339 and lane numbers are identical to those in (A).
The cultural characteristics of L. edodes FMRI0339 and its forty monokaryotic progeny were compared by using two different media, PDA and V8 juice agar plates. Radial growth on plates was assessed by measuring the diameter of the colonies originating from an agar disk (5 mm diameter), taken from the edge of an actively growing colony. Mycelial mass was assessed by harvesting mycelia cultured on a plate overlayed with Miracloth (Calbiochem, La Jolla, CA, USA). Regardless of which strains were cultured, higher growth rate measured by colony diameter was obtained when using PDA. However, mycelial density was increased when using V8 juice agar and at least a 30% increase in the mycelial mass was observed for each strain (data not shown). Thus, V8 juice medium appeared to be better for the enhancement of the mycelial mass. Although there was no significant correlation between virus incidence and growth rate at P < 0.05 by t-test, one out of eight monokaryotic lines showing an active growth rate (> 5 cm) was infected by the mycovirus(Fig. 3). Thus,we tested the virus incidence in actively growing progeny by testing for the presence of virus within the top 20% of actively growing progeny. The virus incidence was reduced significantly, and 2 out of 24 monokaryotic progeny were infected by the virus. However, when we examined the presence of the virus in slow-growing progeny (< 20%), no significant difference from the average was observed. Although the difference in the genetic backgrounds of monokaryotic progeny was obvious due to sexual recombination and further studies of the viral infection on the mushroom production are required, it will be of interest to observe the relatively low incidence of the virus in highly actively growing progeny. The virus-free monokaryotic progeny will provide an opportunity to breed virus-free commercial lines. The establishment of virus-free and -infected isogenic lines is under investigation in order to study the viral effect on mushroom biology.
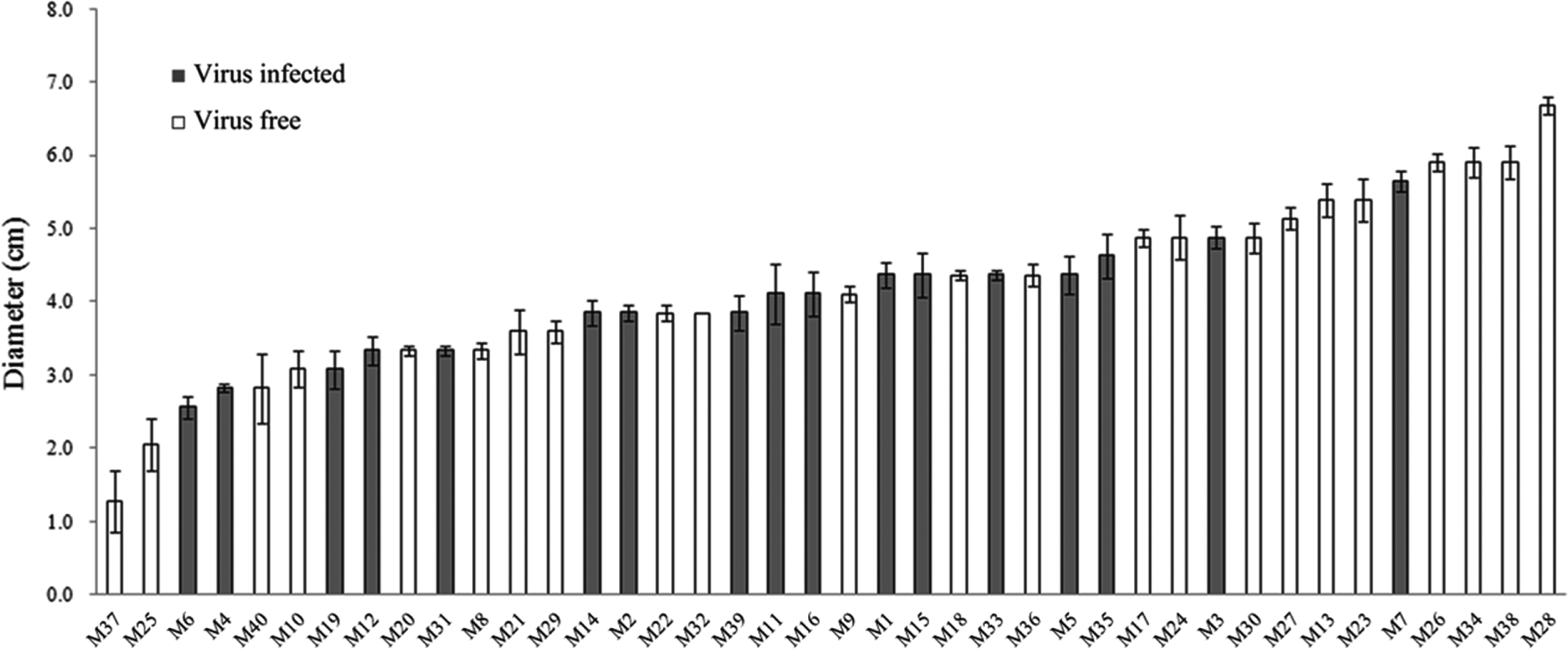
Growth rates of forty monokaryotic progeny. The diameters of colonies after 15 days of culture on PDA were represented in ascending order and strain identification was provided under the x-axis. Error bars represent the standard deviation of three replicates in two independent experiments.
In this study, we described the presence of dsRNA in a popular sawdust-cultivated strain of shiitake mushrooms and suggested the dsRNA as the LeVHKA strain. We observed a surprisingly high incidence of the dsRNA in sexual progeny, suggesting the occurrence of stable meiotic inheritance. In addition, we also showed that the viral incidence decreased in the actively growing progeny, which may suggest an easy way of isolating virus-free progeny.
Acknowledgements
This research was supported by the Bio-industry Technology Development Program, Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea and in part by the KOSEF grant (R11-2008-062-02002-0). This work was in part supported by Korea Research Council of Fundamental Science & Technology (Joint Degree and Research Center for Biorefinery). We thank the Research Center of Bioactive Materials and the Institute for Molecular Biology and Genetics at Chonbuk National University for kindly providing the facilities for this research.