Development of a Model to Predict the Primary Infection Date of Bacterial Spot (Xanthomonas campestris pv. vesicatoria) on Hot Pepper
Article information
Abstract
A population model of bacterial spot caused by Xanthomonas campestris pv. vesicatoria on hot pepper was developed to predict the primary disease infection date. The model estimated the pathogen population on the surface and within the leaf of the host based on the wetness period and temperature. For successful infection, at least 5,000 cells/ml of the bacterial population were required. Also, wind and rain were necessary according to regression analyses of the monitored data. Bacterial spot on the model is initiated when the pathogen population exceeds 1015 cells/g within the leaf. The developed model was validated using 94 assessed samples from 2000 to 2007 obtained from monitored fields. Based on the validation study, the predicted initial infection dates varied based on the year rather than the location. Differences in initial infection dates between the model predictions and the monitored data in the field were minimal. For example, predicted infection dates for 7 locations were within the same month as the actual infection dates, 11 locations were within 1 month of the actual infection, and only 3 locations were more than 2 months apart from the actual infection. The predicted infection dates were mapped from 2009 to 2012; 2011 was the most severe year. Although the model was not sensitive enough to predict disease severity of less than 0.1% in the field, our model predicted bacterial spot severity of 1% or more. Therefore, this model can be applied in the field to determine when bacterial spot control is required.
Bacterial spot, which is caused by Xanthomonas campestris pv. vesicatoria (Xcv), has occurred nationwide on hot peppers in Korea. This disease has become more severe than Cercospora leaf rot (Cercospora capsici) or white leaf spot (Stemphylium solani). At this time, farmers have not instituted control procedures for these diseases because they are not considered sufficiently serious (Kim, 2010). However, an irregular margin of the spot started to develop yellow halos. These severe leaves progressed and ultimately fell. Occasionally, the necrotic spot could be found on fruits, which caused a yield loss. Pathogens overwintered in the diseased leaves could be transmitted by wind and rain (Jones and Pernezny, 2000), as well as seeds. Pathogens can also be found on seedlings during the growth stage of transplants, but they are commonly found in the field after the fruits form (McInnes et al., 1988). Disease control is mainly dependent on fungicides containing copper, and seed disinfection is recommended.
A forecasting model can be applied to properly control disease development. The control, followed by warnings using a forecasting model, can be more effective than periodical control, despite the use of less spray (Kang et al., 2010; Kim and Yun, 2013). De Wolf and Isard (2007) divided the disease developmental stages into overwinter, reproduction, transmission, and pathogenesis. A mechanistic approach to develop a disease forecast model involves analyzing and characterizing the crucial disease stages. Among the commercialized phytobacterial models, meteorological data were applied to examine disease infection (Jacquart-Romaon and Paulin, 1991). The population density was critical to forecast disease development. For example, Erwinia amylovora could infect when the pathogen population reached 105–106 cells/flower, but not less than 103 cells/flower (Billing, 1984). Soybean brown spot occurred when Pseudomonas syringae pv. syringae reached more than 104 cells/g (Cheng et al., 1989). We have developed a bacterial wilt model and found that the population of Ralstonia solanacearum had to reach 109 cell/g of soil to infect the root of hot pepper after overwintering (Kim et al., 2012). These increases in the pathogen population are temperature-dependent. One explanation of why disease development is highly dependent on pathogen density is quorum sensing (Clough et al., 1997). Therefore, population density is an important factor in phytobacterial disease.
In this study, we developed a model of bacterial spot on hot pepper based on meteorological data. The model was designed to predict the initial infection date based on three steps of information: namely, 1) to estimate the dynamic population of the pathogen on hot pepper depending on the moisture conditions of the leaf surface, 2) to determine successful infection of the pathogen from the surface to inside of the leaf, and 3) to determine development of spot symptoms on leaves above a certain population of the pathogen inside the leaf. Each step is required to establish the disease, and the former step must be completed to develop the next step. In addition, the developed model was validated based on collected disease data from 1999 to 2007 at the Rural Development Administration (RDA) monitoring plot in Korea. Each step consisted of empirical models, which were established using the following experiments. The increasing pathogen population was simulated in a liquid culture experiment at varying temperatures, and the decreasing population was simulated by increasing the length of dry periods on the foliar surface. Finally, the minimum density of the pathogen required for disease development and the severity on seedlings depending on the density of injection were examined in pot experiments. In this report, the empirical models required to establish the model are provided.
Materials and Methods
Pathogen population study
A pathogen (No. 11157) of Xcv was obtained from the Korean Agricultural Culture Collection (KACC), RDA Genebank, Suwon, Korea. To select a rifampicin-resistant isolate, the microbe was cultivated for 18 h on Luria Bertani (LB) agar plates with 100 μl/ml rifampicin. To identify rifampicin-resistant pathogens, an isolate that could survive on dried leaves for at least 48 h was selected. The selected Xcv isolate was liquid-cultured in LB solution for 18 h and diluted to 5×106 cells/ml, with an optical density of 0.5 based on spectrophotometry (Biochrome, Cambridge, UK). The culture solution was serially diluted 10,000, 100,000, and 1,000,000 times to make final densities of 5×102, 5×10, and 5×1 cells/ml, respectively. Each dilution (1 ml) was inoculated and shake-cultured in 100 ml of LB liquid at 10°C, 14°C, 18°C, 22°C, 26°C, 30°C, and 34°C at 180 rpm on a shaker. The bacterial suspension (100 μl) was sampled at 0–144 h and was then spread on LB agar plates with rifampicin to count bacterial populations. The colony counting was repeated six or seven times in each temperature regime.
To determine the decreasing population density on dried leaves, Xcv bacterial suspension with OD600 = 0.135 (5×106 cells/ml) was sprayed on a surface of hot pepper leaf until the suspension began to run off. The sprayed seedlings were kept at less than 40% relative humidity (RH) and 24– 27°C for 3 days. The leaves were sampled at 0, 12, 24, 36, 48, 60, and 72 h after spraying. Leaf pieces of 2×2 cm were cut from the center of the foliar samples and vortexed with 2 ml of distilled water. The bacterial density of each sample was counted on rifampicin-LB plates with 6 replications.
Since Xcv bacteria cannot enter cells by themselves, wounds must be generated by wind and rain. Among Xcv on foliar surfaces, only a few epiphytic pathogens can enter successfully. To determine the number of pathogens on leaves that successfully enter the host, the bacterium were artificially injected using a 1 ml syringe (injection) or the leaves were wounded using a needle followed by being sprayed (wounded spray) with a bacterial suspension of OD600 = 0.135. After 12, 24, 36, and 48 h of injection or wounded spraying, the density of the pathogen in the leaf was measured, and symptoms on the leaf were observed for the two artificial infection methods. In addition, the bacterial suspension was diluted until symptoms were not observed and the minimum density required to spot symptoms was determined. The dilutions of the injection were 4, 8, 32, 96, 1,000 and 10,000 times from the original OD600 = 0.135.
Because the growth of Xcv inside the host depends on temperature, and spot requires a specific minimum population, bacterial spot on seedlings could be quantified according to the temperature regimes. Xcv suspension (100 μl) with OD = 0.135 was injected into a leaf of hot pepper seedlings with 5–6 leaves. The densities of the inocula were adjusted to 5×105, 5×104, 104, 5×103, and 5×102 cells/ ml after serial dilutions of 10, 100, 500, 1,000, and 10,000 times, respectively, from OD = 0.135 Xcv liquid culture. The water-soaked margins showing on the inoculated leaves were marked with a pen and then put in a multistep incubator (MTI-201, Eyela, Tokyo, Japan) at 15–35°C with 5°C intervals. Between 7 and 10 days after inoculation, the symptoms of bacterial leaf spot were monitored every day at each temperature and the different initial inocula for 5–7 days. Evaluation of the spot symptoms followed the 0 to 5 scale: 0, no symptoms; 1, spots on the first leaf; 2, most leaves had spots; 3, all leaves had severe spots; 4, leaf death. These scales were assigned a disease severity value: 0 and 1, 0%; 2, 50%; 3 and 4, 100%.
Bacterial spot data in the field were provided by the RDA from 1999 to 2007. Of 101 assessed samples, 43 had a disease incidence of 1% or less. To determine the meteorological variables describing the severity of bacterial spot, a linear regression model (which has the disease severity as Y and various meteorological variables as X) was applied. The model with the highest R2 included temperature, wind speed, and precipitation as X variables. These three variables were selected as the meteorological factors required for Xcv infection.
Outline of the bacterial spot model
Based on the above experiments, models for bacterial spot on hot pepper could be established. The amount of pathogen on a leaf surface increased depending on the wetness period and temperature. At the same time, the amount of pathogen decreased as the length of the dry period increased. The conditions on the surface were conducive to infection if 1) the population density reached the minimum level on the leaf surface and 2) a wound on the leaf caused by wind or rain was present. Once the pathogen infected the leaf, the Xcv population in the leaf decreased by a specific amount. The population of the pathogen inside the leaf started to multiply depending on the temperature and the initial inoculum density. Finally, leaf symptoms showed after reaching a specific Xcv population. Once the calculated population in the model exceeded the threshold, the model could be used to predict the first occurrence date. If the model predicted an earlier occurrence date, then bacterial spot would be more serious during the growing season.
Population increase: Estimating the doubling time for wet leaves
Since Xcv is an epiphytic bacterium, the doubling time (which is the time to double the initial population) depended on temperature. To generate a model equation depending on temperature, regression analysis was performed based on data from liquid culture at 10°C, 14°C, 18°C, 22°C, 26°C, 30°C, and 34°C, and the initial populations were adjusted to 1–10 cells/ml, 10–100 cells/ ml, and 100–1000 cells/ml. The increase in pathogen population at a certain time (t) and initial pathogen populations could be determined as follows:
Population decrease: Estimating the half-life when the initial population decreased to half under dry conditions
Based on experiments on dry leaf surfaces, we could estimate the half-life of Xcv while the population was decreasing. The half-life could be defined as the amount of time before the population reaches half of the initial population density. We assumed that the half-life depends on the duration of the dry period, regardless of temperature. The equation model for half-life is as follows:
Requirements for successful infection of Xcv into the host leaf
In order to sucessfully infect, the pathogens were more 5 × 103 cells/ml as well as the requirements for wind speed and precipitation were met. Based on the regression study between bacterial disease and meteorological data from RDA from 1997 to 2007, a wind speed of more than 3 m/s and precipitation of more than 5 mm/h were required for entering into host leaf (Supplementary data 5). Once the bacterial population entered a leaf, the Xcv population density was decreased to 1/100 times of the epiphytic population, based on the comparison between injection and wound spray study (Supplementary data 3).
Running the developed model on computer
After confirming all required infection processes, the model was run on a PC server in the Epidemiology Laboratory at Seoul National University. Population densities of the pathogen on a host were calculated from one bacterium at the beginning on a leaf. Hourly temperature, relative humidity, and precipitation during the growing season were the input variables for the model simulation. Epiphytic population of Xcv was increased or decreased depending on wetness condition. In order to successfully infect, the pathogens were more 5×103 cells/ml as well as the requirements for wind speed and precipitation were met. The pathogen population inside the leaf was started to colonize from 1/100 times of the epiphytic population at the time of entering. And they were able to build up 1015 cells/ml if temperature of the leaf was appropriate growth. When population of Xcv inside the host reached to 1015 cells/ ml, the model warned the timing of the first infection date which was empirically chosen after running of the model by the Xcv disease and meteorological data from RDA.
Map of the model output of Xcv first infection dates for 2009–2012
The first infection dates were calculated from the model by inputting synoptic observation data, which covered South Korea with 69 areas and were mapped each year from 2009 to 2012. The empty areas were predicted to have no bacterial spot, and the light color represented the area in which the disease was predicted to occur late. Each step of the colored area was 10 days from early July to early September. The area with increasing darkness represented earlier initial observation dates.
Validation of the model outputs compared with the monitored field data for 2000–2007
In total, 94 disease assessment records of bacterial spot on hot pepper were monitored by the RDA at 29 monitoring sites from 2000 to 2007. The 94 monitoring records consisted of 81 diseased data samples (>0.01%) and 11 non-diseased data (0%) samples. Bacterial spot was recorded every month and we assumed that bacterial spot appeared at the end of the month. The synoptic observed meteorological data, which covered 69 sites of South Korea from the Korea Meteorological Administration (KMA) were used. Model outputs were shown as the initial infection dates in each season as the Julian date. To validate model accuracy, the results of the model were compared with the disease records from the monitored fields. After running the model in this study, the 94 results corresponded with the 94 monitored data points and could be divided into the following groups: 1) Bacterial spots were identified in the field and the model predicted the disease (O-O), 2) disease was not found or predicted by the model (X-X), 3) disease was not found but an observation date was predicted by the model (X-O), and 4) disease occurred but was not predicted by the model (O-X). Agreement was calculated based on the following equation:
Differences of the first infection dates between monitored fields and model prediction
Of the 94 monitored data samples from the RDA, 21 results with bacterial spots were monitored in the field and compared with dates predicted by the model (O-O). Each of the 21 results had a monitored and predicted date. Differences between actual and predicted data for the 21 data (O-O) points were categorized as either occurring in the same month, different by 1 month, or different by more than 2 months.
Results
Increase in pathogen population depending on the initial population density and temperature
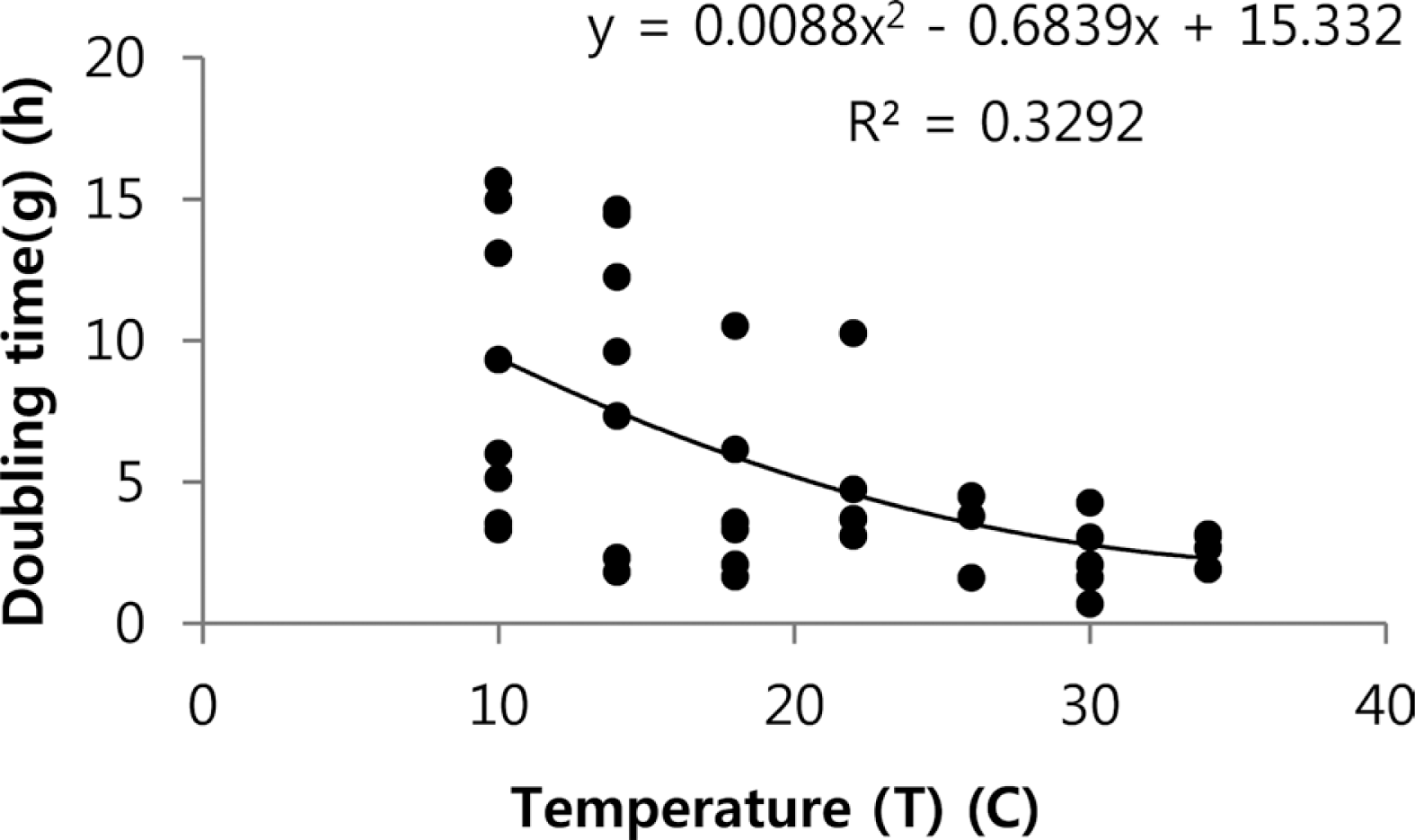
Depending on the initial population at 5, 5×10, and 5×102 cells/ml, as well as the temperature at 10°C, 14°C, 18°C, 22°C, 26°C, 30°C, and 34°C, the shaking cultured growth of Xcv (including exponential and lag phases) varied (Fig. 1). The 5×10 cells/ml of initial density showed a clear bacterial growth pattern depending on the temperature (Fig. 1A). However, 5×102 cells/ml did not experience a lag phase because it started too fast to show differences at different temperatures (Fig. 1B). Also, 5 cells/ml did not grow at 10–18°C because of dilution issues (Fig. 1C). In addition, the exponetial phases at 26–34°C were similar. Since typical growth patterns are required to estimate a doubling time, 5×10 cells/ml of initial inoculum density was chosen. At 10°C, the pathogen survived because the final populations at 144 h were 43 cells/ml. At 14°C, the pathogen only grew to 70 cells/ml by 84 h. However, typical bacterial growth was shown from 18°C to 34°C. In this range, the exponetial phase ranged from 0 to 50 cells/ml, followed by 50 to 300 cells/ml. The duration of the exponetial phase after inoculation in liquid LB depended on the incubation temperatures (Fig. 1C). For example, exponetial phase at 18°C were 48–60 h, at 22°C were 36–60 h, and at 26°C, 30°C, and 34°C were 12–24 h. Since doubling time in each temperature was calculated (Supplementary data 1), we could plot temperature as the x-axis and doubling time (g) as the y-axis. The best fitted regression equation was second order, as shown in Fig. 2. The doubling time of the Xcv pathogen at a certain temperature could be estimated from the following equation:
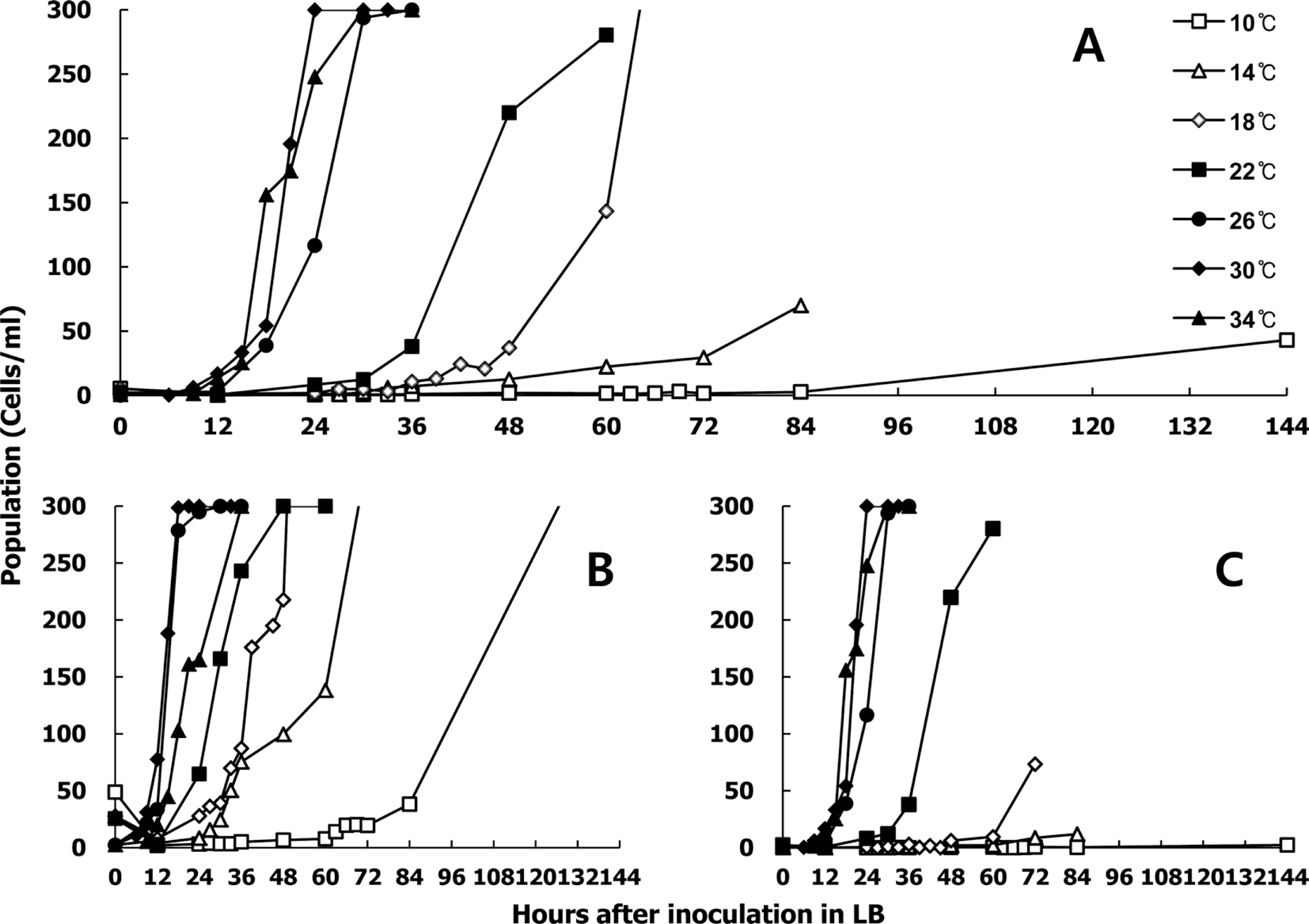
Growth of rifampicin-resistant Xanthomonas campestris pv. vesicatoria in liquid culture. Initial inoculum density was adjusted to OD600 = 0.135 and then diluted to final densities of (A) 5×10 cells/ml, (B) 5×102 cells/ml, and (C) 5 cells/ml. The seven temperature regimes were 10°C, 14°C, 18°C, 22°C, 26°C, 30°C, and 34°C.
Decrease in the pathogen population depending on the length of the dry periods of the host leaf
As the dry period increased, the population of Xcv decreased significantly. Only 1/100 of the initial population survived after 12 h of dry period. Although we sprayed 5×106 cells/ml, less than 100 cells/ml survived at 24 h and less than 10 cells/ml survived after 48 h of dry period. Among six replications, only one bacterium on the leaf surface survived after 72 h of dry period (Fig. 3). Regardless of the temperatures, we obtained an average of 24 data points (Supplementary data 2).
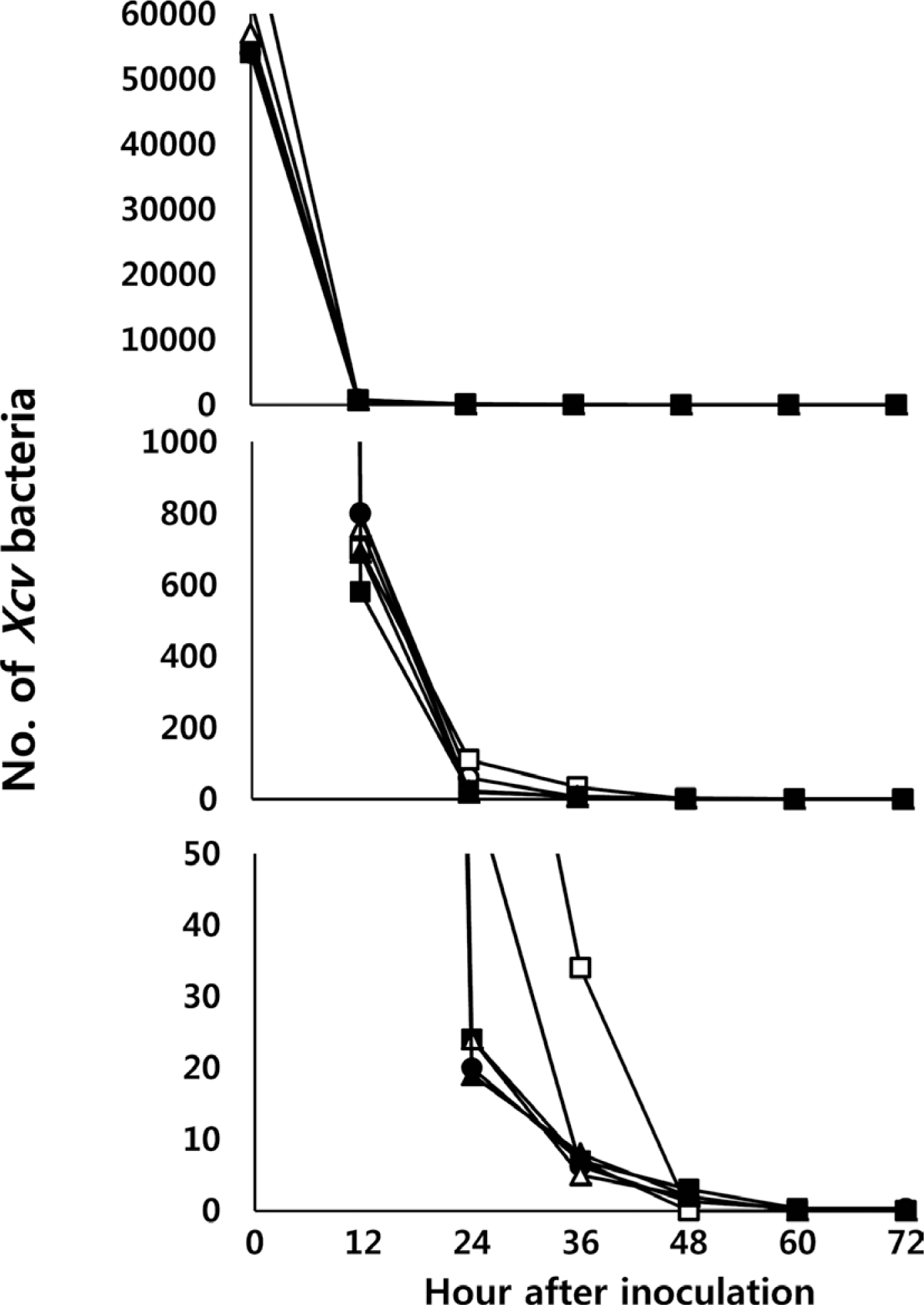
Decrease in Xanthomonas campestris pv. vesicatoria (Xcv) populations 24, 36, 48, 60, and 72 h after dry periods on the surface of a leaf. Each symbol represents one replication. Total six replications were conducted. From top to bottom panels, the scales of the bacterial population are decreasing. The treatments of dryness were increased to 72 h.
Pathogen populations in the host leaf by injection or through artificial wounds
Compared with injection or wound inoculation of Xcv with OD600 = 0.135, the measured populations between the two methods were similar at 12 h. However, the populations by means of injection were about 100 times higher than those using wounds at 36 and 48 h (Supplementary data 3). Serial dilution from OD600 = 0.135 were evaluated to identify the required minimal concentration to show the bacterial spots on leaves, and up to a hundredfold diluted samples could still show symptoms. However, more than hundredfold dilution showed no symptoms on leaves (Supplementary data 4).
Spot symptoms depending on various temperatures and initial populations
The bacterial spot symptoms on the host leaf were observed from 5 to 8 days after Xcv infection, and the intensity of the symptoms depended on the temperature regimes and initial inocula (Fig. 4). At 5×106 cells/ml for the initial population, all inoculated leaves showed typical spot symptoms after 5 days of inoculation (excluding at 15°C). At 5×105 cells/ml of initial population, all inoculated leaves showed spots after 6 days of inoculation (excluding at 15°C). At an initial population of 5×104 cells/ml, all inoculated leaves showed spots after 7 days of inoculation (excluding at 15°C). At an initial population of 104 cells/ml, only 67% of inoculated leaves at 30°C and 25°C showed symptoms, while 56% of the inoculated leaves at 35°C showed symptoms. A total of 22% of leaves at 20°C showed spots. However, we did not identify any symptoms at 5×103 cells/ml (excluding at 30°C, with 11% showing symptoms). Based on these experiments, we determined the lowest population level inside the leaves that can induce symptoms. In other words, symptoms appeared regardless of the temperatures if the initial population inside the leaves was 5×104 cells/ml or more. However, symptoms at initial populations of 104 cells/ml depended on the treated temperatures. Also, the initial density of 5×103 was too low to induce symptoms on host leaves.
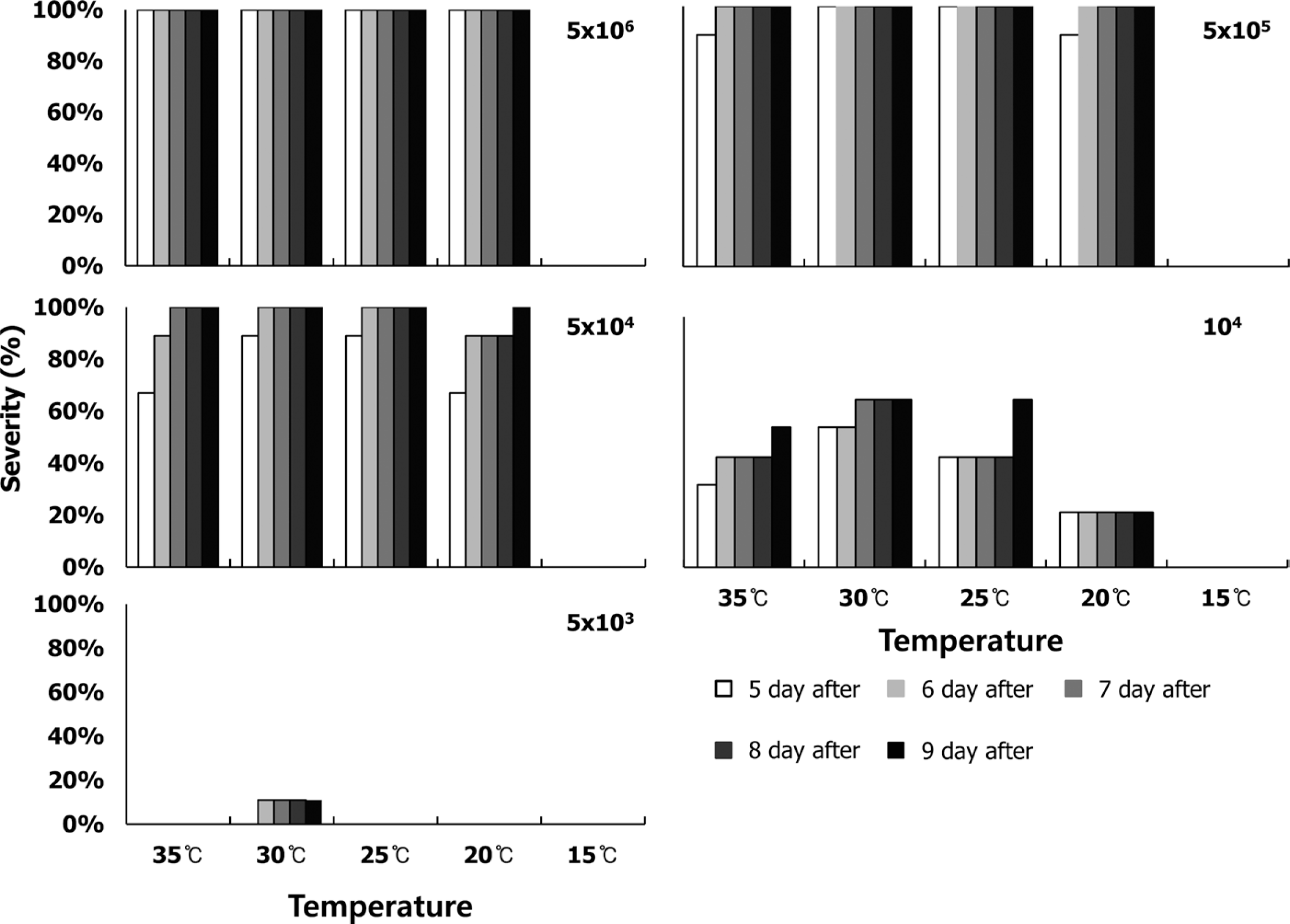
Severity of bacterial spots on the leaves of hot pepper seedlings according to the five initial populations: (A) 5×106, (B) 5×105, (C) 5×104, (D) 104, and (E) 5×103 cells/ml and depending on the five temperatures (15°C, 20°C, 25°C, 30°C, and 35°C). Severity (%) was the percent of diseased leaves and symptoms observed at 5, 6, 7, 8, and 9 days after inoculation.
Identifying necessary environmental factors for infection based on the monitored field data
By performing simple regression analysis of severity (response variables) and various meteorological data (explanatory variables), we chose several regression models with high R2 and included temperature, precipitation, and wind speed variables. The best describing model of the incidence of bacterial spot was within temperature ranges of 21–23°C, more than 1 m/s wind speeds, and more than 25 mm of precipitation (R2 = 0.29, Supplementary data 5).
Outline of the bacterial spot model
We can now simulate the dynamics of Xcv populations on the surface of and inside pepper leaf. Depending on the temperature and dry period, the population of Xcv on the surface of pepper seedlings changed significantly. To successfully enter and infect the host leaf, the Xcv population on the leaf surface required more than 5×103 cells/ml. In addition, wind speeds greater than 3 m/s and precipitation greater than 4 mm/h were required to form wounds on the host leaves (Supplementary data 5). According to the injection study on pepper seedlings, the population density of Xcv would be diluted by 10−2 when entering the leaf. Since the Xcv population required more than 104 cells/ml to induce symptoms, the surface population should be at least 106 cells/ml. In this model, warning signs began at a minimum of 1015 cells/ml (threshold) in host tissue (Fig. 5). The first occurrence date of bacterial spot in the model was programmed to be more than the threshold of the pathogen population (Fig. 5). Therefore, the bacterial spot model could estimate the first occurrence date depending on the population dynamics on the surface of and inside the host tissue.
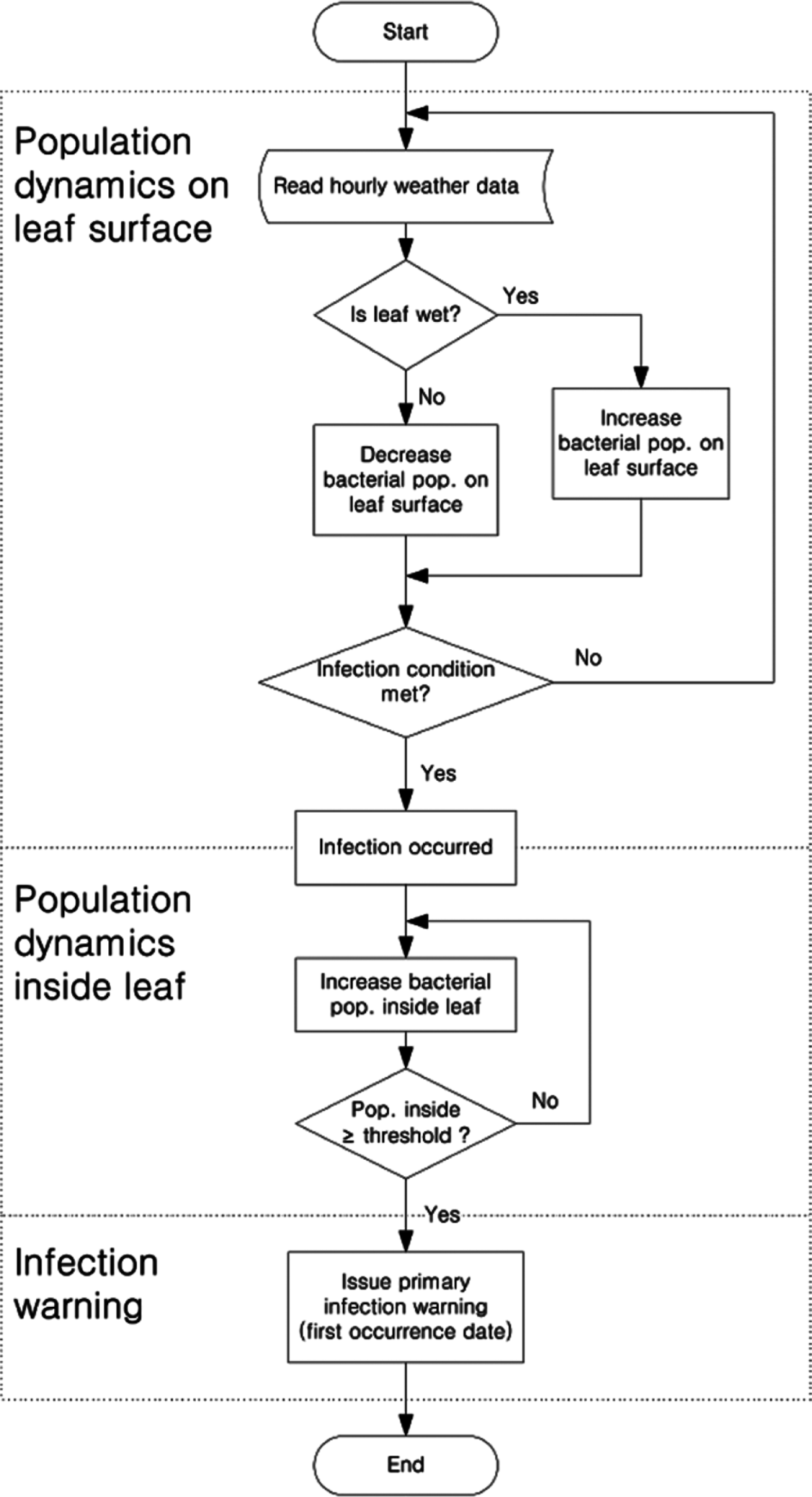
Flowchart of a forecast model for bacterial spot caused by Xanthomonas campestris pv. vesicatoria on hot pepper. The pathogen population on the surface of leaves increased or decreased depending on wetness. If rain levels exceeded 4 mm/h and wind speed was greater than 3 m/s, wounds formed on the leaves. Also, if the pathogen population reached 5,000 cells/g, infection was successful. Once they could infect inside the leaf, their population decreased by a hundredfold from the infected population and would subsequently grow depending on the temperature. Ultimately, the initial infection date was decided when the population was over 1015 cells/g.
Map of the model output of the first infection dates for 2009–2012
From 2009 to 2012, the model predicted that the most severe year was 2011, with approximately 50% disease incidence. At that time, the west coast and middle of the Korean Peninsula was diseased from early or mid-July. The years 2009 and 2010 were predicted to be less severe than the other years, and most of the diseased areas were in western areas. On the other hand, eastern areas, including Kangwon and Gyeongbuk (primary production areas of hot pepper), were at lower risk (Fig. 6).
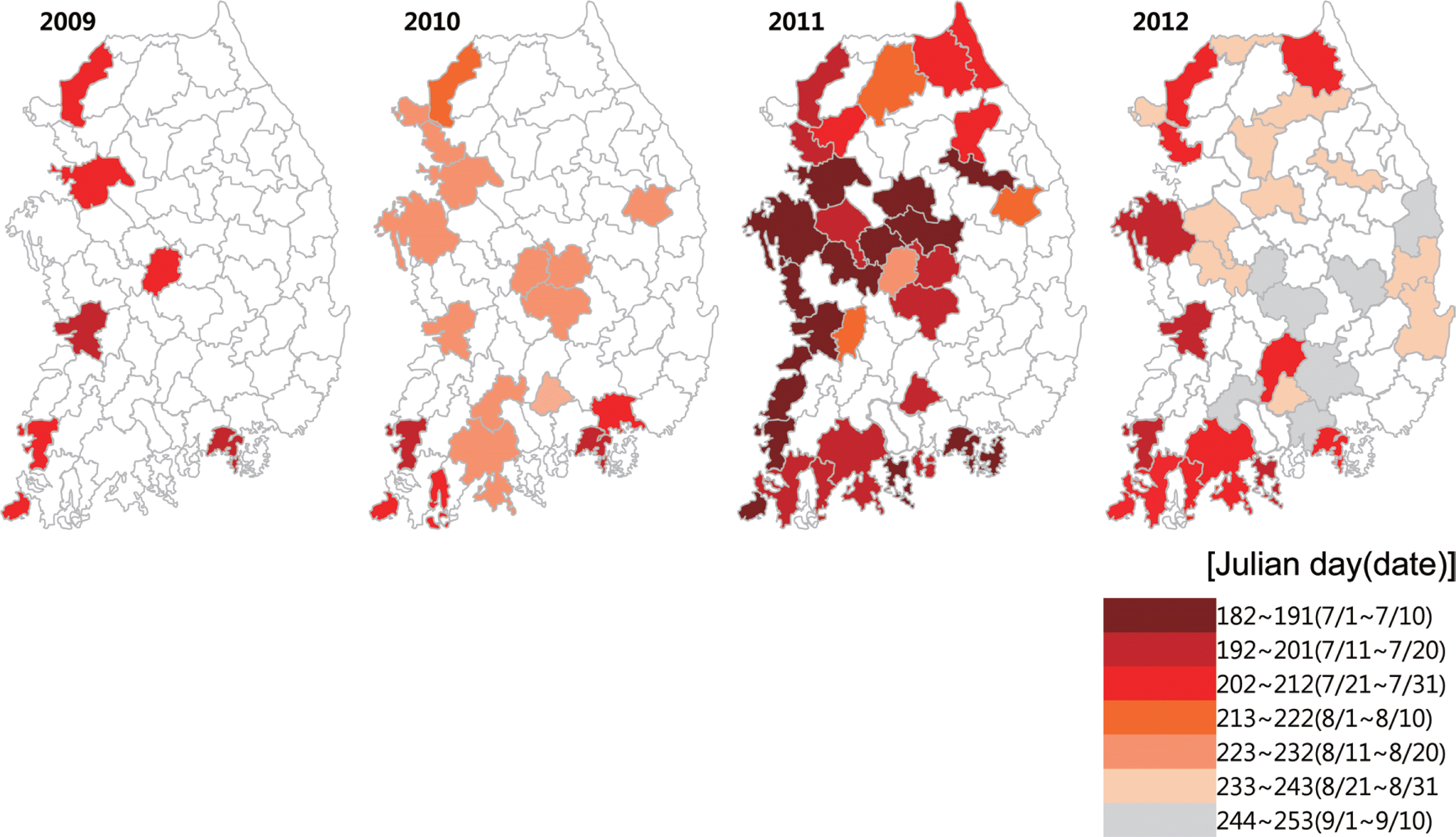
The model map of the predicted initial infection dates of bacterial spot on hot peppers from 2009 to 2012. The developed bacterial spot model was based on meteorological data from the Korea Meteorological Administration (KMA). The model was based on synoptic observing data, which cover 69 sites of South Korea. The colored area represents the initial infection dates of bacterial spots from early July to early September.
Validation of the model using monitoring records from 2000 to 2007
The model developed in this study was operated corresponding to the 94 assessments from 24 monitoring fields from 2000 to 2007. Among the results of 94 operating fields by the model, only 25 were at risk for bacterial spot. Of the 25 at-risk fields, 21 had bacterial spots and 4 did not. Of the 69 low-risk areas, 9 were non-diseased in the field. Therefore, our developed model accurately predicted the disease results for 30 areas. Then, the model had a 31.9% accuracy compared with observed disease areas (Fig. 7 and Supplementary data 6).
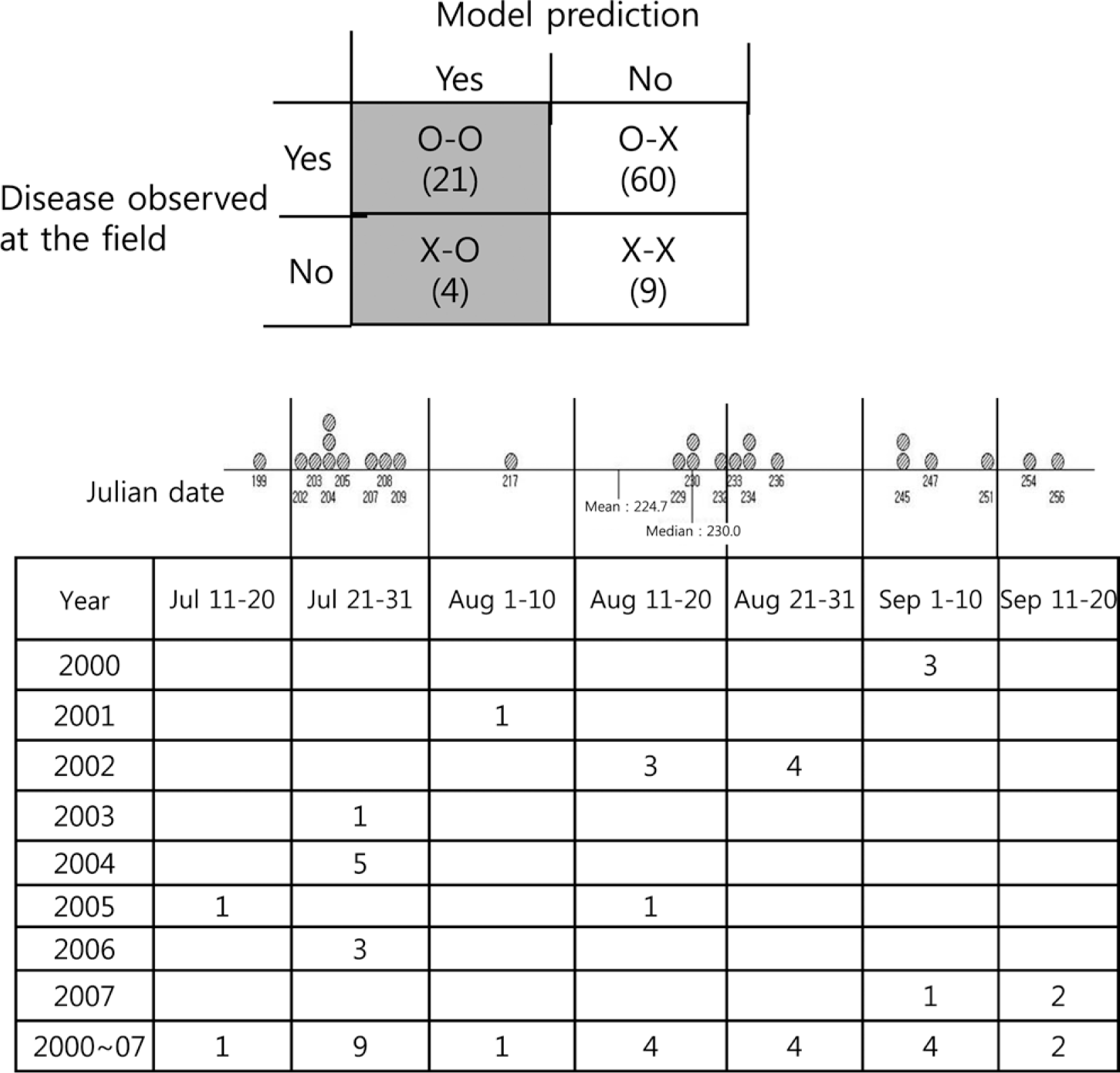
Analysis of the 25 initial infection dates from the model predicted the bacterial spot from 2000 to 2007 at the monitoring sites, which included 29 fields. Among the 25 model predicted initial infection dates, 21 fields contained observed bacterial spots and 4 fields did not contain disease. The below table shows the 25 predicted dates according to the year and Julian date.
Differences in the first infection dates between the model and observed fields
Differences in the first occurrence dates between the 21 results examined for bacterial spots in the field and predicted to be at-risk based on the model are shown in Fig. 8. Among the 21 results, 7 were observed and predicted to be within the same month, 11 differed by 1 month, and 3 differed by more than 2 months. Among the observed data, incidence of the disease at Seosan in September 2009 was 15.7%. This suggested that bacterial spot had started earlier, but was measured more recently (Fig. 8). In addition, the model warned of the disease in mid- or late August, but the disease was observed in September at Boeun, Gongju, and Cheongsong in 2002. These differences were mainly due to late monitoring in the field. However, the model warned of the disease in early September, but the disease had already occurred in August at a 1% incidence in Andong, Uiseong, and Cheongsong in 2007. These were issues associated with the model forecasting (Fig. 8).
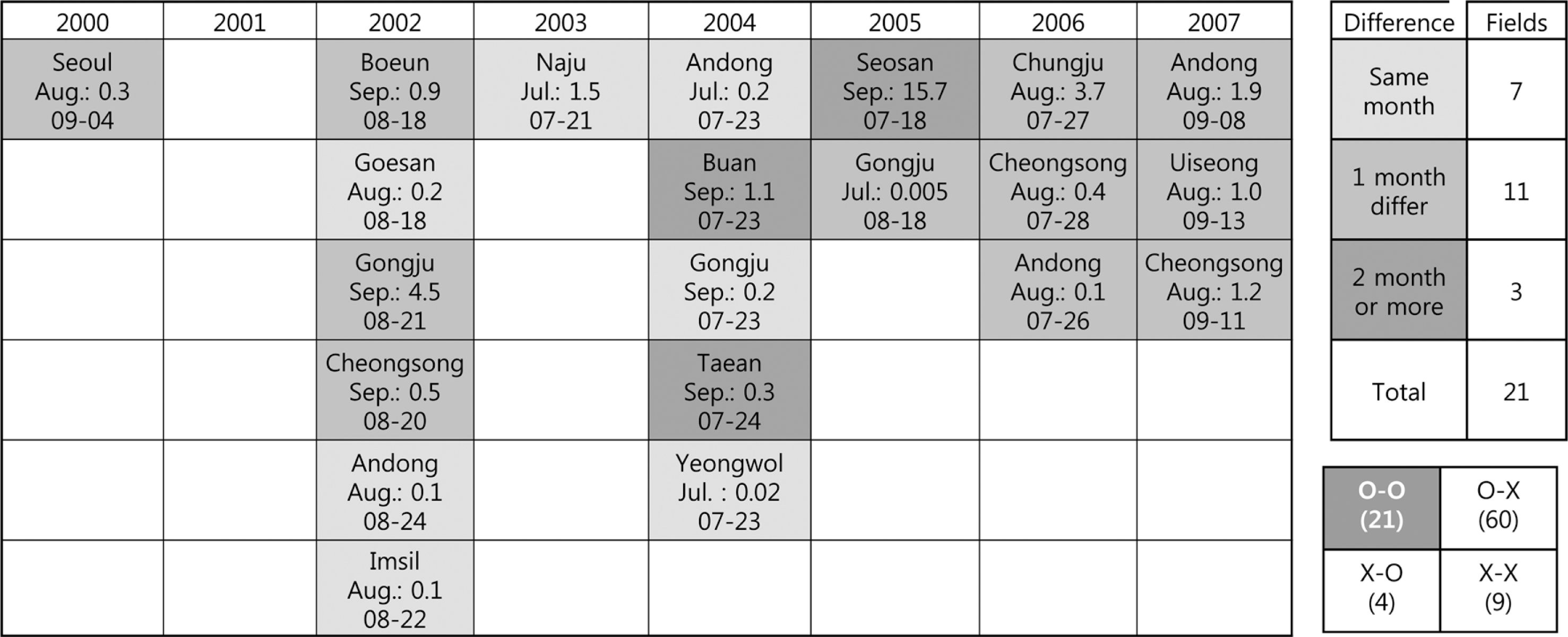
Differences in the initial infection dates between monitored fields and model predicted bacterial spots from 2000 to 2007 in Korea. A forecasting model was operated by the synoptic observation data consisting of 75 sites from the Korea Meteorological Administration (KMA). Differences between observed and predicted infection dates were categorized as either occurring in the same month, 1 month difference, or 2 month difference, represented by darker areas.
Discussion
The model in this study was used to monitor pathogen population dynamics depending on temperature and humidity of the host surface. If the infection conditions, that is, the density of the pathogen and wound are associated, then the pathogen would grow in the host depending on the temperature inside the leaves. Finally, bacterial spot on the leaves would form when the density of the pathogen population reached a specific level. De Wolf (2007) used the mechanistic modeling approach to characterize overwintering, reproduction, transmission, and pathogenicity to develop a disease prediction model. Numerous prediction models have been generated since 1990, but the majority have been used to identify infection substages and reproduction substages. Our developed model has been applied to identify infection conditions necessary to colonize leaves and for pathogenesis.
The empirical model can be used to identify necessary conditions for disease infection based on the correlation coefficient of regression analyses between monitored disease assessment data as independent variables and various meteorological data as dependent variables. Our findings of Xcv infectious conditions are not sufficient to describe the amount of bacterial spot based on meteorological variables, such as temperature, wind, and rain. When we considered temperature and precipitation without wind speed, the best model’s R2 value was 0.41. This was significantly higher than 0.29 of the selected model in our study considering temperature, precipitation, and wind speed. Since wind is required for wound formation and bacterial disease, our infection model used these three meteorological conditions, including wound formation.
We had been decided that the critical threshold of the infection was 1015 cells/ml which was unrealistically high and had never been reached at the laboratory study. When the developed model was run by the meteorological data coincident with Xcv data from RDA, the population on computer was empirically increased upto 1015 cells/ml or more and it was easily accomplished under conducive condition. According to the study of Xcv bacteria entering into leaf either by injection or wound (Supplementary data 3), epiphytic pathogen by entering through wound was decreased to 1/100 times by injection. It was speculated that Xcv population inside the leaf was deceased from epiphytic population due to natural wound caused by wind. Since the pathogen population on a leaf decreased to 1/100 after natural wound infection, at least 5×103 cells/ml or more on a leaf was necessary to successfully colonize. It was necessary to maintain the environmental condition inside the host leaf after successful infection, then they were built up 1015 cells/ml to show the spot symptom on leaf. Finally, our developed model tells us the infection moment as the first infection date when the pathogen population inside a leaf increased to the threshold.
Agreement of the model results compared with the monitored assessments from the field reached a maximum of 31.9%. The majority of the monitored assessments in the field were not severe during 2000–2007. Of the 94 data points, 11 were 0% infected and 34 were 0.01–0.1% infected. If we consider that no-disease was 0.1% instead of 0%, our model would reach 55% agreement (Supplementary data 6–1). Actually, less than 0.1% severity of bacterial spot on leaves was not severe enough to warrant control during harvest. When we consider validation of the model, it is important to forecast the proper time to spray fungicide rather and exclude a mild disease that does not require control. In addition, mild symptoms of bacterial spot on the leaves do not affect yield; thus, the farmer would not require control. Overall, our model accuracy could reach 55% when excluding mild diseases (<0.1% severity of Xcv) in the fields. Also, our model can forecast more than 1% disease severity, which may warrant chemical control by farmers.
According to comparisons of the first observation dates between model predictions and field assessments, the differences among years were larger than among locations within a year. For example, the first observation dates in 2003, 2004, and 2006 were much earlier than those in 2000–2002 and 2007, regardless of the location. Bacterial spot is more severe in the early typhoon season than late typhoon season (Kim, 2010), which suggests that bacterial spot in Korea varies more significantly by year than by location. It is important to consider that the model forecasting may overlook mild infection within a field. Also, the model may be late in predicting the timing of infection, such as in Andong, Euisung, and Cheongsong in 2007, in which bacterial spots were already observed in August 2007, but the model forecasted them for September. Excluding these incorrect predictions, no other issues occurred using the model. Although the model is not sensitive enough to predict less than 0.1% disease severity, it is sufficient to predict occurrence levels that would require control of the disease in advance. In addition, the model can be applied practically in the field.
Although we were not shown the results (K. H. Lee, personal communication), comparisons between field data and model outputs at Chungbuk Province for 2010–2012 showed that the model accurately forecasted bacterial spot. For example, bacterial spot in 2011 was most severe in Chungbuk Province fields (about 10%), and the model predicted bacterial spot at three locations in early July and one location in late July in 2011. On the other hand, at Chungbuk, the disease incidence was 2% in 2010 and 0% in 2012. The model forecasted the disease in 2010 at only one location in early August. Also, the model forecasted the disease in late August or early September in 2012, which did not require control. The model in Chungbuk Province for 2010–2012 was validated and could be used to predict the disease for farmers to prepare the fields.
Compared with the pattern of bacterial wilt on hot pepper in our previous studies (Kim et al., 2012), bacterial wilt developed early in the season and the damaged area was wider than the bacterial spot. In addition, bacterial wilt initiated from the aboveground part of the plant, which is more serious than starting on the leaf. At this time, it is difficult to identify typical symptoms of bacterial spot on fruits in Korean fields. Therefore, it is not necessary to control the disease at locations where the disease is not present or where the first observation dates were late August or September.
Based on our results on chili pepper (Shin and Yun, 2010) and its changes after infection by Phytophthora blight, anthracnose, and bacterial wilt or spot, as well as the effect of climate change, bacterial spot may become much more common and could become severe. Thus, protective measures should be taken in the future. In addition, the two bacterial diseases (spot and wilt) are not easily controlled by conventional methods. Although these bacterial diseases have not significantly reduced pepper yield (until now), it is necessary to monitor these bacterial diseases and prevent them.
Acknowledgements
This study was performed with the support of the Cooperative Research Program for Agricultural Science & Technology Development (Project No. PJ007395), Republic of Korea.