Genetic Mapping of a Resistance Locus to Phytophthora sojae in the Korean Soybean Cultivar Daewon
Article information
Abstract
Phytophthora root and stem rot reduce soybean yields worldwide. The use of R-gene type resistance is currently crucial for protecting soybean production. The present study aimed to identify the genomic location of a gene conferring resistance to Phytophthora sojae isolate 2457 in the recombinant inbred line population developed by a cross of Daepung × Daewon. Single-marker analysis identified 20 single nucleotide polymorphisms associated with resistance to the P. sojae isolate 2457, which explained ~67% of phenotypic variance. Daewon contributed a resistance allele for the locus. This region is a well-known location for Rps1 and Rps7. The present study is the first, however, to identify an Rps gene locus from a major soybean variety cultivated in South Korea. Linkage analysis also identified a 573 kb region on chromosome 3 with high significance (logarithm of odds = 13.7). This genomic region was not further narrowed down due to lack of recombinants within the interval. Based on the latest soybean genome, ten leucine-rich repeat coding genes and four serine/threonine protein kinase-coding genes are annotated in this region, which all are well-known types of genes for conferring disease resistance in crops. These genes would be candidates for molecular characterization of the resistance in further studies. The identified R-gene locus would be useful in developing P. sojae resistant varieties in the future. The results of the present study provide foundational knowledge for researchers who are interested in soybean-P. sojae interaction.
Phytophthora sojae Kauffmann and Gerdemann is an oomycete pathogen that causes destructive root and stem rot in susceptible soybean plants [Glycine max (L.) Merr.]. From 1996 to 2014, about 25 million tons were lost annually due to Phytophthora root and stem rot (PRR), with an average annual loss of more than 1.1 million tons (Allen et al., 2017; Wrather and Koenning, 2006, 2009; Wrather et al., 2001).
In wet soil, zoospores of P. sojae are attracted by exudates released from soybean roots. After reaching a root surface, the zoospores attach and transform into a cyst that germinates to produce a hypha that penetrates the plant within a few hours of inoculation (Morris and Ward, 1992). P. sojae colonizes root and stem tissues as reproducing oospores. Seedling damping-off occurs at early stages of susceptible varieties, whereas root and stem rot, leaf yellowing, and wilting typically develop at later stages of growth followed by plant death (Schmitthenner, 1985).
Soybean varieties carrying an effective resistance (R) gene for defending against P. sojae isolates carrying an avirulence (Avr) gene rapidly develop a hypersensitive response within hours after infection (Staskawicz et al., 1995). R-gene resistance can provide effective protection from P. sojae and is widely used in developing resistant varieties in several major soybean growing countries such as the US, China, Brazil, and Japan (Dorrance, 2018). Jang and Lee summarized a comprehensive list of Rps (resistance to P. sojae) documented in previous studies. To date, 34 allele have been mapped to nine loci: Rps1a, 1b, 1c, 1d, 1k, 7, 9, UN1, YU25, YD29, HN, Q, WY, HC18, X, and Rps gene in the Waseshiroge soybean cultivar on chromosome 3 (Cheng et al., 2017; Demirbas et al., 2001; Li et al., 2017; Lin et al., 2013; Niu et al., 2017; Sugimoto et al., 2007, 2011; Sun et al., 2011; Weng et al., 2001; Wu et al., 2011a; Zhang et al., 2013b; Zhang et al., 2013b, 2019); Rps3a, 3b, 3c, 8, and SN10 on chromosome 13 (Burnham et al., 2003; Gordon et al., 2006, 2007; Yu et al., 2010); Rps4, 5, 6, 12, and JS on chromosome 18 (Gordon et al., 2007; Sahoo et al., 2017; Sandhu et al., 2004; Sun et al., 2014); Rps2 and UN2 on chromosome 16 (Demirbas et al., 2001; Huang et al., 2016; Lin et al., 2013); RpsZS18 on chromosome 2 (Yao et al., 2010); Rps11 on chromosome 7 (Ping et al., 2015); RpsSU on chromosome 10 (Wu et al., 2011b); Rps10 on chromosome 17 (Zhang et al., 2013a); and RpsYB30 on chromosome 19 (Zhu et al., 2007).
PRR was first identified two decades ago in South Korea (Jee et al., 1998). PRR began to be seriously considered as an issue as soybean cultivation in paddy soil increased (Ministry of Agriculture, Food and Rural Affairs, 2019). Paddy soils are usually wet because of poor drainage. This condition usually facilitates infection by soil-borne pathogens such as P. sojae or Fusarium spp. (Dorrance, 2018; Schmitthenner, 1985). Especially, a few-week period after soybean planting, there is heavy and frequent rain in June to July; 163 and 289 mm in 30-year average (1981-2010) according to the Annual Climatological Report of Korea (available at http://book.kma.go.kr/ebook/ebView.asp?eb_no=202006301610443121&r=R), which would make favorable condition for the plant pathogens. As the use of paddy fields for soybean production is increasing year by year, PRR might occur more frequent and severe near future.
PRR has been little studied in S. Korea and interaction between soybean and P. sojae in domestic soybean varieties is largely unknown. Kang et al. (2019) examined the presence of Rps resistance against four isolates in 20 universal soybean cultivars widely planted in South Korea. Surprisingly, susceptibility was observed in 14-19 of the 20 cultivars. About 50% of the evaluated cultivars were susceptible. Only a few cultivars, such as Daewon, showed resistance to isolates, implying that most cultivars are vulnerable to P. sojae. Still, few reports on interactions between domestic soybean varieties and P. sojae isolates in S. Korea are available. In the present study, the universal cultivar, Daewon, is first characterized for resistance to P. sojae via linkage analysis using a recombinant inbred line (RIL) population that segregates in reactions to P. sojae isolate 2457.
Materials and Methods
Plant materials.
The mapping population was developed from crosses between female parent Daepung and male parent Daewon (Kim, 2018). Briefly, the first cross for F1 seeds occurred in 2012. Eleven F1 seeds were planted in the winter of 2012, and 103 F2 seeds were collected. Generations of seeds were produced by single-seed descent from F2 to F5 by 2015. Seeds of F5-derived RILs were increased at the university farm of Chungnam National University in 2018. The two parents, Daepung and Daewon, have white flowers, determinate growth habit, and yellow seeds. Daepung and Daewon were recently reported as susceptible and resistant to the P. sojae isolate 2457, respectively (Kang et al., 2019). The isolate 2457 was isolated from the cultivar Uram in Gyeongbuk province in 2016 (Kang et al., 2019).
Evaluation of resistance to P. sojae.
The two parents and RILs of the mapping population were examined for resistance to P. sojae using hypocotyl inoculation (Dorrance et al., 2004). Briefly, 10 to 15 seedlings were grown in a 13 cm pot filled with soil. An inoculum was prepared on a clarified V8 medium about 10 days before inoculation. V8 medium fully covered by P. sojae mycelia was macerated twice using a 50 ml syringe and transferred into a 10 ml syringe. Seven days after sowing, a 1 cm slit was made on hypocotyl of the seedlings, and 0.2 to 0.4 ml of a mycelial slurry was placed in the slit. The inoculated seedlings were placed in dark and humid condition for 16 h. Seedlings were then incubated in a growth chamber (14 h [light]/10 h [dark], temperature 25°C, and humidity >70%) and their reactions were observed 7 days after inoculation. The reaction of each RIL was determined as a percentage of the number of killed to the number of seedlings. Seedlings were resistant (R) if <20% died, susceptible (S) if >80% died, or intermediate (I) if 20 to 80% died. This experiment was replicated three times.
DNA extraction and single nucleotide polymorphism (SNP) genotyping.
Genomic DNA of parents and RILs was extracted using cetyl trimethyl ammonium bromide as described by Doyle and Doyle (1987). Samples were genotyped with Axiom 180 K SoyaSNP arrays (Affymetrix, Santa Clara, CA, USA) (Lee et al., 2015). SNP data were preprocessed as follows. Beginning with 169,028 SNPs obtained from the array, 143,083 undefined or nonpolymorphic SNPs between the two parents were removed. Additional 184 and 335 SNPs were eliminated because of ≥10% missing values and ≥10% heterozygosity, respectively. Two RILs were also excluded because of ≥10% missing values.
Goodness-of-fit test and single-marker analysis of variance.
A goodness-of-fit to Mendelian segregation ratios was calculated using chi-square (χ2) analysis to examine patterns of the phenotypes in mapping populations. Marker-trait association was analyzed via a single-marker analysis of variance (ANOVA) using the lme4 package (Bates et al., 2015) in R (R Core Team, 2019). Bonferroni correction (P < 1.00E-6) was applied to determine the most stringent threshold to minimize false positiveness in ANOVA results.
Construction of genetic map and linkage analysis.
A genetic map of Daepung × Daewon population was constructed using a software IciMapping version 4.1 (Meng et al., 2015). First, the function BIN (i.e., Binning) was used to remove redundant markers. Remaining markers were grouped and ordered using the function MAP (i.e., mapping). Linkage analysis then conducted using the function BIP (i.e., Biparental mapping), to confirm marker– trait associations identified by single-marker ANOVA. For quantitative analysis, disease score (i.e., R or S) of an RIL was converted to quantitative form as an average percentage of dead seedlings over three replicates. Additivity was calculated as an allelic effect of Daepung relative to Daewon.
Results
Phenotypic assay for resistance to P. sojae.
Two parents and 71 RILs of the Daewon × Daepung population were evaluated for R-gene type resistance to P. sojae isolate 2457. Daewon and 35 RILs were resistant, whereas Daepung and 24 RILs were susceptible (Table 1, Fig. 1). Four 4 RILs exhibited intermediate resistance with 41-59% dead, and eight RILs were indeterminant because of inconsistent results; consequently, 12 RILs were excluded from analyses.

Goodness-of-fit test for segregation ratios of recombinant inbred lines in the Daepung × Daewon population following inoculation with Phytophthora sojae isolate 2457

Results from parents, Daepung (left) and Daewon (right), observed 7 days after the inoculation of Phytophthora sojae isolate 2457.
A goodness-of-fit test demonstrated that segregation ratios were consistent with the Mendelian segregation, suggesting that the trait is qualitatively controlled by a single gene (χ2 = 1.49, P = 0.36). Distributions of phenotypes were discrete with high peaks at both ends, typical of single-gene inheritance.
Single-marker ANOVA.
A total of 25,761 SNPs was analyzed to find significant marker–trait associations. ANOVA identified 20 highly significant SNPs located in an 859 kb region on chromosome 3, for which P-values ranged from 1.01E-7 to 1.68E-11 (Table 2). The estimated R2 (%) ranged from 57 to 67, indicating that the locus would be a major gene controlling the resistance (Table 2).
Genetic mapping of a locus associated with resistance to P. sojae.
An alternative analysis was used to confirm strong associations of the genomic region with resistance to P. sojae. Over 25,000 SNPs were reduced to 6,364 SNPs by binning, which subsequently were used to construct genetic maps (Table 3). A total of 263 SNPs were integrated on chromosome 3 with the total genetic length of 87.3 cM (Table 3).
Linkage analysis narrowed down to a 573 kb region (3,893,390 to 4,752,969 bp) from the 859 kb interval identified from the single-marker ANOVA, where 46 genes are annotated based on the latest soybean reference genome Glyma2 (http://soybase.org). The 573 kb region on chromosome 3 was detected with high significance (logarithm of odds = 13.7), explaining 66.4% of phenotypic variance (Table 4, Fig. 2). The positive additive effect indicates that the percent of dead seedlings was higher in RILs with the Daepung allele than in those with the Daewon allele for identified SNPs, indicating that Daepung provides susceptibility alleles (Table 4).
A genomic region identified for resistance to Phytophthora sojae isolate 2457 in the Daepung × Daewon RIL population

Graphical presentation of Rps locus associated with resistance to Phytophthora sojae isolate 2457 in the Daepung × Daewon recombinant inbred line population. The genetic map is presented with a plot of logarithm of odds (LOD) score from linkage analysis with quantitative scales. Hatched lines on the LOD plots indicate the LOD threshold. The 1-and 2-LOD intervals are displayed as black bars and solid lines, respectively.
Discussion
In the present study, Phenotypic variation among RILs in the Daepung × Daewon RIL population was evaluated for R-gene mediated resistance to P. sojae isolate 2457. A genomic region significantly associated with the resistance, containing multiple Rps genes, was identified via single-marker ANOVA and linkage analysis. This report is the first to characterize a major soybean variety cultivated in S. Korea for the resistance to P. sojae.
Single-marker ANOVA and linkage analysis identified a locus with high levels of statistical significance on chromosome 3 (Table 3, Fig. 2). Since no other region was identified from ANOVA and linkage analysis, resistance is apparently conditioned on the expression of a gene or a few in this single locus. The identified region partially overlaps or is in close proximity to genomic regions where over ten Rps alleles were reported, including five Rps1 alleles—Rps1a, Rps1b, Rps1c, Rps1d, and Rps1k—and Rps7, Rps9, RpsHC18, RpsHN, and Rps of cv. Waseshiroge, RpsQ, RpsUN1, RpsYD29, RpsYU25, RpsWY, and RpsX (Fig. 3) (Cheng et al., 2017; Demirbas et al., 2001; Li et al., 2017; Lin et al., 2013; Niu et al., 2017; Niu et al., 2017, 2011; Sun et al., 2011; Weng et al., 2001; Wu et al., 2011a; Zhang et al., 2013b; Zhong et al., 2017, 2019). Since no RILs display recombination within the ~573 kb marker interval, the target resistance locus could not be further narrowed down in the genome. The Rps of Waseshiroge, RpsYD29, RpsHN, and RpsHC18 mostly overlap in the identified interval of the present study (Fig. 3). An Rps gene locus of the cv. Waseshiroge mapped between Satt009 and T0003044871 (3,931,955-4,486,048 bp) (Sugimoto et al., 2011) that largely overlaps the interval identified in the present study, whereas RpsYD29 (Zhang et al., 2013b) and RpsHC18 (Zhong et al., 2017) are located at the ends of the interval, flanked between SattWM82-50 and Satt1k4b (3,857,715-4,062,474 bp) and between BARCSOYSSR_03_0269 and BARCSOYSSR_03_0272 (4,514,938-4,611,259 bp), respectively.
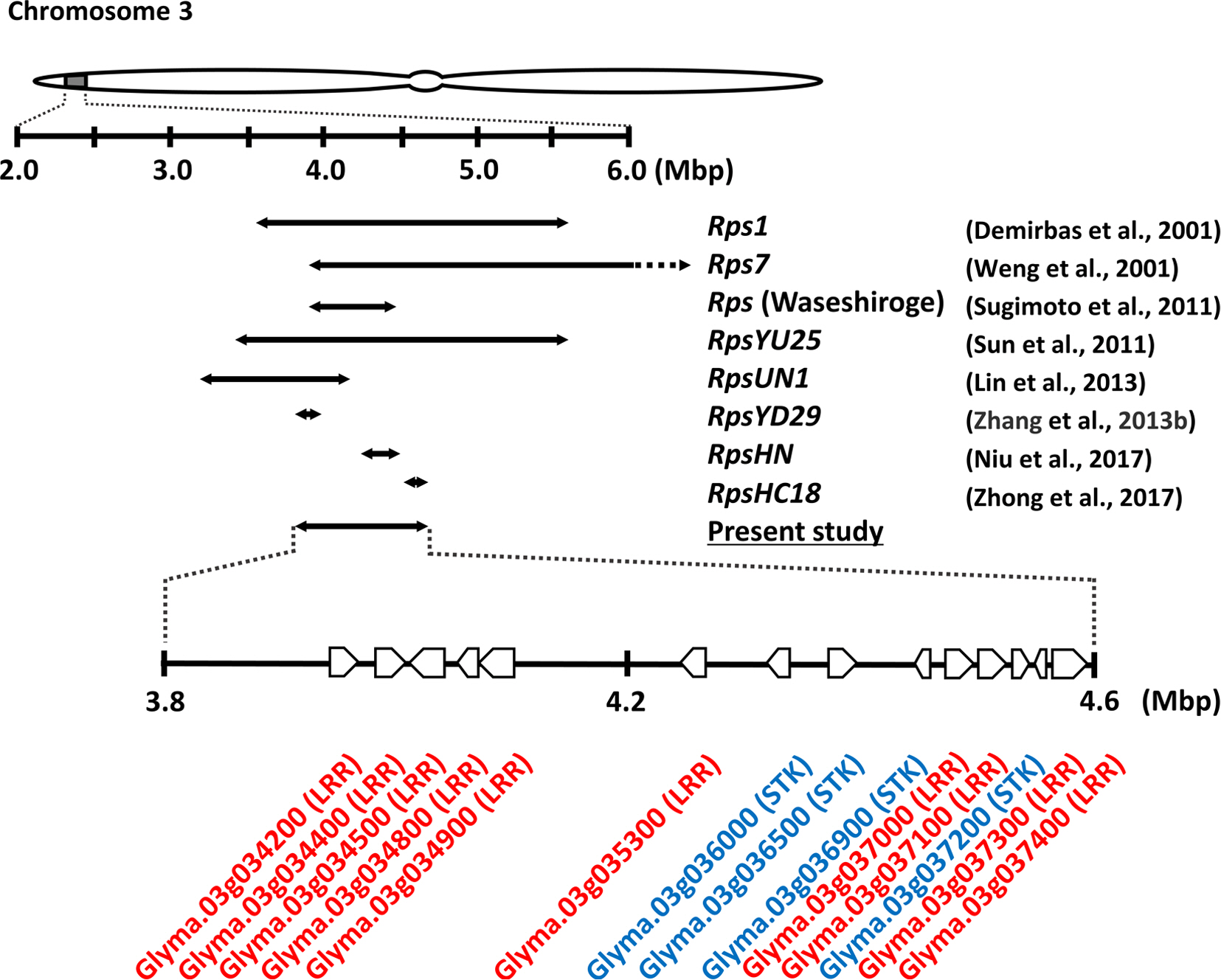
A schematic diagram of the identified Rps region and annotated gene models (Glyma.Wm82.a2.v1). Selective known Rps genes overlapping this locus are presented by arrows, indicating marker intervals of them as previously reported. The dashed part of the interval presents outside of 6.0 Mbp. Ten leucine rich repeat-and four serine/threonine protein kinase-coding genes annotated in this genomic region (3.8 to 4.6 Mbp) are displayed at the bottom.
The gene encoding nucleotide-binding site-leucine rich repeat (NBS-LRR) protein is a well-known category of resistance gene in plants. In soybean–P. sojae interactions, Rps1k is best characterized and codes for a type of NBSLRR protein (Gao and Bhattacharyya, 2008). Additionally, previous studies of soybean–P. sojae interactions report genes encoding NBS-LRR protein as a candidate gene (Li et al., 2017; Niu et al., 2017; Zhang et al., 2013b; Zhong et al., 2017). The identified locus was further examined with the latest gene annotation, i.e., Glyma.Wm82.a2.v1, at Soybase (http://soybase.org) (accessed September 1, 2020). A total of ten LRR type genes were found in the genomic region (3,893390-4,642,893 bp), including Glyma.03g034200, Glyma.03g034400, Glyma.03g034500, Glyma.03g034800, Glyma.03g034900, Glyma.03g035300, Glyma.03g037000, Glyma.03g037100, Glyma.03g037300, and Glyma.03g037400. These genes would be primary candidate genes for future studies of resistance to P. sojae isolate 2457 in Daewon.
Serine/threonine protein kinase (STK) gene is another type of plant resistance gene (Zhong et al., 2019). Several STK genes are responsible for disease resistance in plants, including Xa21, Xa21D, and Xa3/Xa26 for bacterial blight in rice; Lr10 for wheat leaf rust; and Rvi12_Cd5 for apple scab (Feuillet et al., 1997; Padmarasu et al., 2018; Song et al., 1995; Sun et al., 2004; Wang et al., 1998). In soy-bean–P. sojae interactions, Zhang et al. (2013a) mapped Rps10 on chromosome 17 and reported Glyma17g28950 and Glyma17g28970 (i.e., Glyma.17g196200 and Glyma.17g192300 in Glyma.Wm82.a2.v1, respectively) as candidate genes for Rps10. These two genes were predicted to encode plant-type Ser/Thr protein kinase, suggesting that the mechanism of resistance mediated by Rps10 might differ from Rps1, Rps2, and Rps4 that represent NBSLRR genes. Four STK-coding genes are annotated in the identified genomic region of the present study, namely, Glyma.03g036000, Glyma.03g036500, Glyma.03g036900, and Glyma.03g037200. These are also strong candidate genes for resistance to P. sojae isolate 2457 and should be further examined.
In summary, Daewon was confirmed as a genetic source to P. sojae isolate 2457 and the 573 kb region of chromosome 3 was identified via linkage analysis. There were at least 14 annotated genes associated with disease resistance found in this interval, thus these genes in Daewon will need to be investigated to advance our understanding in further study. It is also unknown whether the resistance allele of Daewon is a novel gene or an alternative form of Rps1, Rps7, or other Rps genes previously reported in the several resistance sources. Investigation of allelic variation or relationship in this locus among the resistance sources will be of interest in further work. This genomic region can be used to improve resistance to P. sojae of major soybean varieties in breeding programs in S. Korea and the understanding from the present study will provide fundamental knowledges for future studies of molecular interaction between soybean and P. sojae.
Acknowledgments
The present study was funded by the Next-Generation Bio-Green 21 Program (Project No. PJ01333701; Title: Identification of resistance to Phytophthora sojae in the Korean soybean core collection and NAM population), Rural Development Administration, South Korea.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.

