Diversity in Betasatellites Associated with Cotton Leaf Curl Disease During Source-To-Sink Movement Through a Resistant Host
Article information
Abstract
Cotton leaf curl is devastating disease of cotton characterized by leaf curling, vein darkening and enations. The disease symptoms are induced by DNA satellite known as Cotton leaf curl Multan betasatellite (CLCuMuB), dominant betasatellite in cotton but another betasatellite known as Chili leaf curl betasatellite (ChLCB) is also found associated with the disease. Grafting experiment was performed to determine if host plant resistance is determinant of dominant population of betasatellite in cotton (several distinct strains of CLCuMuB are associated with the disease). Infected scion of Gossypium hirsutum collected from field (the source) was grafted on G. arboreum, a diploid cotton species, resistant to the disease. A healthy scion of G. hirsutum (sink) was grafted at the top of G. arboreum to determine the movement of virus/betasatellite to upper susceptible scion of G. hirsutum. Symptoms of disease appeared in the upper scion and presence of virus/betasatellite in the upper scion was confirmed via molecular techniques, showing that virus/betasatellite was able to move to upper scion through resistant G. arboreum. However, no symptoms appeared on G. arboreum. Betasatelites were cloned and sequenced from lower scion, upper scion and G. arboreum which show that the lower scion contained both CLCuMuB and ChLCB, however only ChLCB was found in G. arboreum. The upper scion contained CLCuMuB with a deletion of 78 nucleotides (nt) in the non-coding region between A-rich sequence and βC1 gene and insertion of 27 nt in the middle of βC1 ORF. This study may help in investigating molecular basis of resistance in G. arboreum.
Begomovirus belonging to the family Geminiviridae is the largest genus, with viruses causing diseases in many economically important plants. Genome of begomoviruses consist of either one or two circular ssDNA molecules of ~ 2.5–3.1 kb. The two components of bipartite begomoviruses are referred to as DNA A and DNA B and both are, for the most species, essential for symptomatic infection of plants. Monopartite begomoviruses are often associated with DNA satellites known as alphasatellites and betasatellites. These begomovirus/betasatellite/alphasatellite complexes are widespread in the Old World (OW) and represent the largest group of begomoviruses (Briddon et al., 2004). The disease complex is transmitted by a single species of whitefly, Bemisia tabaci.
Monopartite begomoviruses may require betasatellite to induce disease symptoms in their hosts (Mansoor et al., 2006). Betasatellites are dependent on the helper virus for their replication and share no sequence identity to their helper begomoviruses (Briddon and Stanley, 2006). They are capable of interacting with multiple begomoviruses (Mansoor et al., 2003). Betasatellites have an adenine (A) rich region, a βC1 gene and a highly conserved sequence of about 100 nucleotides known as satellite conserved region (SCR). These complexes are widespread throughout the OW and cause severe disease losses to several crops (Amin et al., 2006).
Cotton leaf curl disease (CLCuD) complex that causes severe losses to cotton crop in the Indian subcontinent is associated with several monopartite begomoviruses (9 different begomoviruses have been identified) and a single species of betasatellite known as Cotton leaf curl Multan betasatellite (CLCuMuB) (Akhtar et al., 2015). In late 1990’s several cotton varieties resistant to the disease were introduced but resistance was broken at the end of the 20th century. Since then the resistance breaking, recombinant begomovirus, Cotton leaf curl Burewala virus (CLCuBuV) has been the only begomovirus associated with the disease in Pakistan (Akhtar et al., 2015; Siddique et al., 2015). This recombinant virus is associated with a betasatellite which is also recombinant. This recombinant betasatellite has derived most of its sequences from the original CLCuMuB but a small portion within the SCR is derived from tomato leaf curl betasatellite. Changes in the dominant begomoviruses and their associated satellites have been shown in the last decade which may be due to the selection pressure exerted by host resistance (Amrao et al., 2010). Thus, host plant resistance seems to be an important determinant of selection of dominant begomovirus complex.
Plants are constantly exposed to a large number of pathogens, which can result in severe damage to the plant. To overcome the problem plants have evolved various types of resistance mechanisms e.g. non-host resistance (NHR) mechanism (Yun et al., 2003). Resistance against plant viruses often involves restriction on virus movement from initially infected cells (Bian et al., 2006). Once the virus is introduced into the plant and it gains control of the plant resistance, the next step is the movement from cell to cell to reach all parts of the plant. Plant viruses use cytoplasmic bridges called plasmodesmata (small tunnels) for short distance movement while long distance movement is accomplished through phloem tubes (Jeske, 2009). In addition, geminiviruses also have to pass through nuclear pores in both directions. This process is carried out by the nuclear localization and nuclear export signals proteins involved in virus movement (Jeske, 2009).
Two virus proteins are involved in begomovirus movement; one facilitate the nuclear shuttling and binds to the viral DNA while the other opens the gate of plasmodesmata and promote the transfer of the viral DNA from one cell to the other cell. In monopartite begomviruses the coat protein (CP/AV1) acts as nuclear shuttling protein (NSP) while the pre coat protein (PCP/AV2) acts as the gate keeper. For bipartite begomoviruses DNA-B encodes BV1/NSP protein for nuclear shuttling and moment protein (MP/BC1) for plasmodesmatal transfer (Wege and Pohl, 2007; Noueiry et al., 1994). Betasatellite associated with monopartite begomoviruses also plays a role in the movement of the virus by facilitating viral DNA amplification and to assist it to reach to all parts of the plant (Saeed et al., 2007). The βC1 protein also increases the replication of the helper begomovirus and overcome the host defense mechanisms (Saeed et al., 2007; Iqbal et al., 2012).
The genus Gossypium contains approximately 50 different cotton species. Of these only four species produce fiber and are grown commercially. These include the two diploid species (G. arboreum and G. herbacium) with an Asian origin, and the two tetraploid species (G. hirsutum and G. barbadancae) of American origin (Ullah et al., 2014). G. hirsutum has now replaced the other three cotton species and is grown globally, producing more than 90% of the world total fiber products. However, it is facing certain challenges which include the problems of attack from pests and diseases (Siddique et al., 2014). The G. arboreum, a native genotype of cotton in the Indian sub-continent has a poor yield and fiber quality but is completely resistant to abiotic and biotic stresses (Akhtar et al., 2010). Begomoviruses are phloem-limited and transmitted by insect vectors but cannot be seed and sap transmitted (Liu et al., 1999). Currently no efforts have been successful to detect CLCuD in G. arboreum in the field (Ullah et al., 2014). Under the present study a grafting assay was developed to analyze the transmission efficiencies of helper virus and betasatellite variants of cotton leaf curl disease complex between susceptible and resistant cotton varieties. The purpose of graft-inoculation was to provide a direct contact between the phloem of scions (both susceptible and infected scions) and root stocks of G. arboreum plants.
Two cotton varieties belonging to G. arboreum (Ravi) and G. hirsutum (CIM-496) were graft inoculated using bottle shoot grafting technique (Akhtar et al., 2013). The plants were grafted in such a way that G. arboreum acted as root stock (bridge) and G. hirsutum as a scions. Lower scion (source) was taken from G. hirsutum field infected with Cotton leaf curl Multan virus (CLCuMuV) and has been grafted on G. arboreum. A healthy scion of susceptible variety of G. hirsutum (sink) was grafted at the top of G. arboreum. The double graft-inoculated plants along with the healthy/non-graft-inoculated plants were kept in the same green house, with an ensured provision of optimum growth conditions and insect free environment. The grafting experiment was repeated twice and a total of 12 plants were grafted each time.
A total of 30 samples from graft-inoculated plants (that include fresh plant leaves from plants acting as source, G. arboreum acting as bridge and susceptible scion acting as sink) and healthy control (negative control) plants were collected after 30 and 85 days for the screening of viral components. Total genomic DNA was isolated from the plant leaves by Cetyl Trimethyl Ammonium Bromide (CTAB) method (Doyle and Doyle, 1990) after each grafting experiment.
Twenty ng/μl dilution of DNA was prepared and subjected to RCA using Phi-29 DNA polymerase (Haible et al., 2006). RCA amplified DNA was subjected to PCR using universal primers Beta01/Beta02 (Briddon et al., 2002) and Begomo-F/Begomo-R (Amrao et al., 2010) for the amplification of betasatellite and begomovirus, respectively. PCR amplified DNA was purified and cloned into the pTZ57R/T vector (Fermentas). The full length cloned products for the betasatellite were confirmed with KpnI restriction enzyme while the clones for begomovirus DNA component were confirmed with BamHI, HindIII, EcoRI and MluI restriction endonucleases. The confirmed cloned products were sequenced commercially (Macrogen, Korea). The sequenced data were subjected to basic local alignment search tool (BLAST) and finally compared with already available sequences in the database. Phylogenetic analysis was done by making phylogenetic tree being rooted on Cotton leaf curl Multan Alphasatellite (CLCuMuA).
Southern blot hybridization was used to detect the presence and accumulation level (titer) of betasatellite in the graft-inoculated plants. Total DNA extracted from G. hirsutum infected with the cotton leaf curl viruses was used as a positive control and healthy G. hirsutum (with no infection) was used as a negative control. Probe for CLCuMuB was prepared using Dioxiginen (DIG) (Roche) DNA labeling kit and a reaction mixture of 25 μl was prepared. DIG PCR mixture comprised of 5pg-10pg template DNA, 2.5 μl 10X Taq polymerase buffer (Fermentas), 5 μl of 2 mM DIG dNTPs, 2.5 mM MgCl2, 0.75 μM each of primers, and 1.25 units Taq DNA polymerase (Fermentas). Approximately equal amount of genomic DNA (10 ug) were loaded in each well, electrophoresed on 1% (w/v) agarose gels, blotted onto positively charged Hybond-Nylon membranes (Roche) and UV cross-linked. Probes of virus fragment (virion-sense) and full length betasatellite were PCR amplified using specific primers CLCV1/CLCV2 (CCGTGCTGCTGCCCCCATTGTCCGCGTCAC/CTGCCACAACCATGGATTCACGCACAGGG) and universal primers Beta01/Beta02 (Briddon et al., 2002), respectively. Hybridization was performed at 55°C for 12–15 hrs followed by stringency washing at the same temperature. Hybridization signals were detected on X-ray film after treatment with CDP-Star (Roche). Hybridization experiment was repeated twice for all the DNA samples collected after each grafting experiment.
Success of grafting was 100% as all the grafted scions were survived till the end of experiment. Typical symptoms of CLCuD were observed in the upper scion after 85–90 days of grafting; however, no such disease symptoms could be seen in G. arboreum. However, the mild symptomatic evidence of CLCuD in G. arboreum (variety Ravi) plants through grafting using resistance breaking recombinant begomovirus, CLCuBuV was recently reported by Akhtar et al. (2013). RCA of the DNA samples collected after 30 days of grafting showed positive amplification for CLCuMuV and betasatellites in the infected lower scion (source) but no such amplification was observed in susceptible upper scion (sink) and G. arboreum (the bridge). Diagnostic amplification (RCA) was positive for DNA extracted (after 85 days of grafting) from infected scion, susceptible scion and root stock samples. This amplified DNA when subjected to PCR for the amplification of begomovirus/betasatellite, positive amplification for both betasatellites and begomovirus was seen in upper susceptible scion (sink) along with the lower infected scions (source) however, only (ChLCuB) was amplified from the root stock of G. arboreum. The presence of begomovirus/betasatellites in the upper scion suggests that long distance transport of helper virus and betasatellites via the phloem occurs in G. arboreum. The cotton variety G. arboreum seems to be not fully resistant since it allows at least the viral trafficking via phloem tissues (Akhtar et al., 2013). This may occur just in rare cases, but even this low efficiency of transfer from helper virus and betasatellites seems sufficient to result in infection of the healthy scion of the susceptible host G. hirsutum. All plants were kept under aseptic conditions in glass house so no chance for the viral vector/contamination could exist and hence the presence of betasatellite in the upper scion and G. arboreum is merely from the infected scion.
Hybridization signals for the betasatellite were seen in all the samples collected from lower infected and upper susceptible scions while no such hybridization signals were detected in any of the sample collected from G. arboreum root stock (Fig. 1). DNA extracted from infected G. hirsutum scions is shown in lanes number 3, 7 and 12, susceptible G. hirsutum scion samples are presented in lanes 6, 9, & 14 while G. arboreum root stock samples are shown in lanes 4, 5, 8, 10, 11 & 13 (Fig. 1). The lanes 4 and 10 contain DNA of G. arboreum root stock that were closely located to infected scion, and lanes 5 and 11 contain DNA of G. arboreum root stock slightly far away from the infected scion.
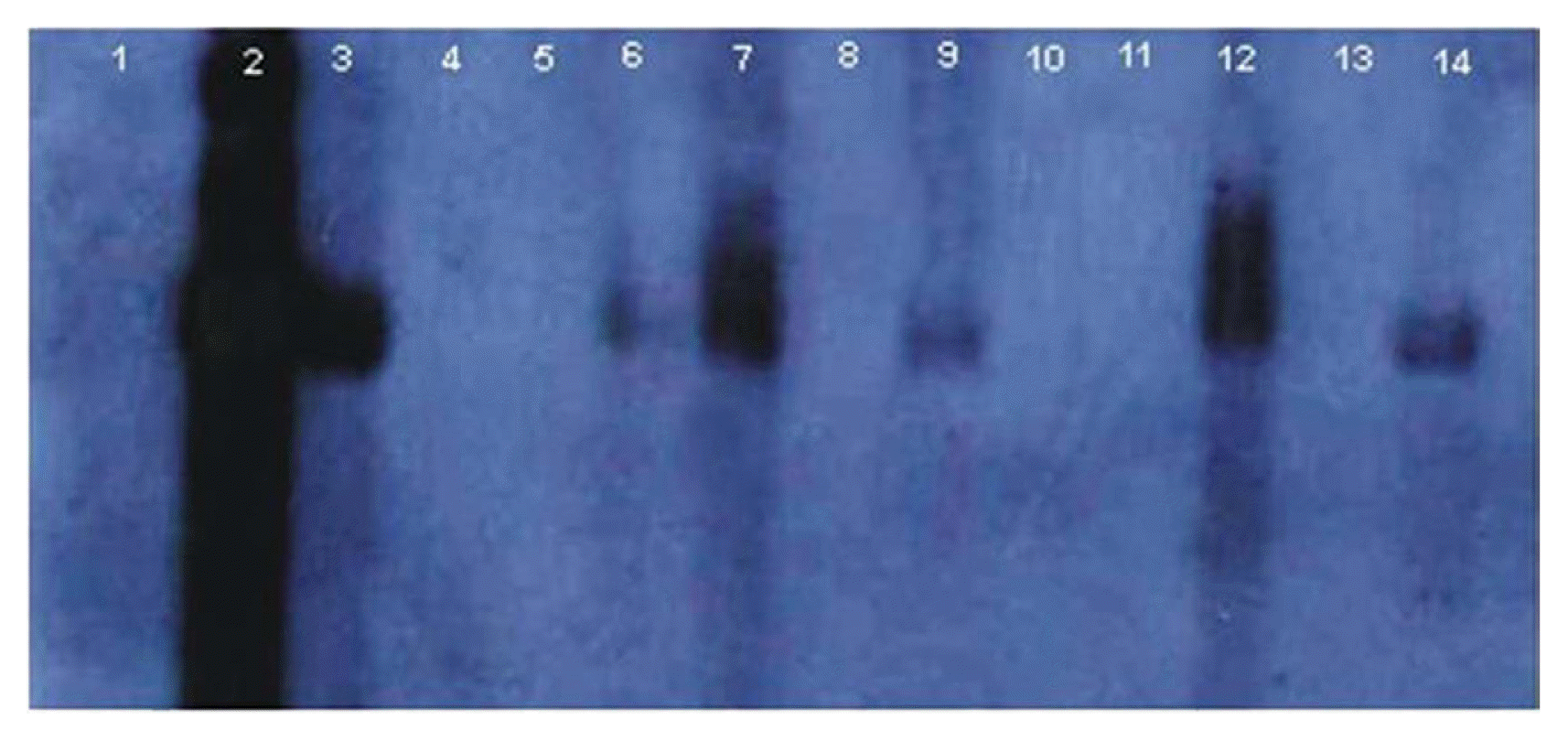
Southern hybridization for the detection of betasatellite using Cotton leaf curl Multan betasatellite (CLCuMuB) sequence as a probe. Negative control [genomic DNA of healthy cotton plant (lane 1)], positive control [genomic DNA of infected cotton plant (lane 2)], infected scions of grafted plants (lanes 3, 7 & 12), root stocks of double grafted plants (lanes 4, 5, 8, 10, 11 & 13) and susceptible scions of grafted plants (lanes 6, 9 & 14).
The betasatellites cloned from infected scions show 100% sequence identity to CLCuMuB while those cloned from susceptible scions show 96% sequence identity to CLCuMuB. The betasatellites cloned from G. arboreum rootstock show 100% sequence identity to ChLCB (Table 1). Phylogenetic tree constructed (rooted on CLCuMA sequence) further confirms that betasatellites cloned from infected and susceptible scions show close similarity to CLCuMuB while the betasatellites cloned from G. arboreum closely resemble ChLCB (Fig. 2). Same pattern was seen for all the samples collected from all the 12 plants.

Phylogenetic dendrograms based upon the alignments of complete nucleotide sequences of betasatellite isolated from G. hirsutum infected scion (scion 1), susceptible scion (scion 2) and G. arboreum root stock (stock) and betasatellite sequences collected from national center for biotechnology information (NCBI). Vertical distances are arbitrary. Horizontal distances are proportional to genetic distances. Phylogeny was rooted on CLCuMA. Numbers at the nodes represent the percent of times at which branching were supported. Betasatellites used for the comparison are Cotton leaf curl betasatellite (CLCuB), Cotton leaf curl Multan betasatellite (CLCuMuB) and Chilli leaf curl betasatellite (ChLCB).
The sequence alignment of betasatellites cloned from infected and susceptible scions show that the betasatellites existing in the susceptible scions have a deletion of 78 nt in the non-coding region between A-rich sequence and βC1 gene, and result in the addition of 27 nt in the middle of βC1 open reading frame (ORF) (Fig. 3). This addition of 27 nucleotides did not change the βC1 ORF but it may result an accumulation of nine amino acids to the βC1 protein.
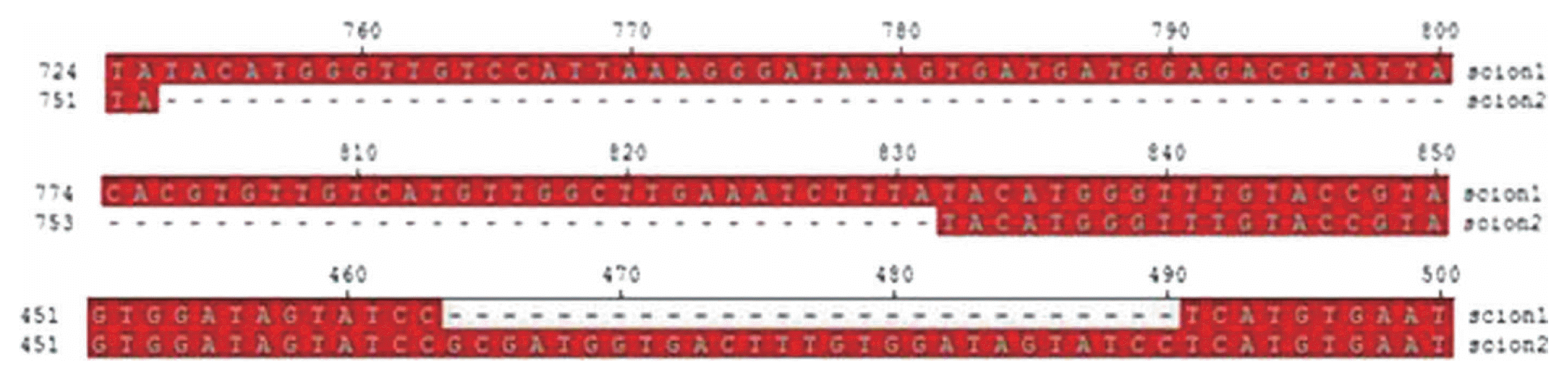
An alignment of sequence of betasatellite cloned from infected scion (scion 1) and susceptible scion (scion 2).
Sequence analysis of betasatellites revealed that the betasatellites cloned from susceptible scions have a close similarity to CLCuMuB. Interestingly, mutations were detected in the non-coding region and also within the βC1 coding region of DNA of the CLCuMuB during source to sink moment through G. arboreum. These variants of CLCuMuB may have enabled the replication and movement of the virus and induction of disease symptoms in the upper scion by breaching the resistance of G. arboreum. The presence of CLCuMuB in the upper scion indicates that the late induction of disease symptoms in the upper susceptible scion might be due to the movement/replication of betasatellite population from the lower infected scion to the upper healthy susceptible scion (Ullah et al., 2014).
The betasatellites cloned from the G. arboreum root stock show close similarity to ChLCB. Once ChLCB moved to G. arboreum, the plant may have provided favorable conditions to overcome CLCuMuB and hence it could become the prominent betasatellite. This may suggest that Rep protein of helper virus present in fewer amounts may have supported the replication of ChLCB in G. arboreum. However, we are unable to confirm this possibility due to our inability to detect low levels of begomovirus in root stock.
Sequence data of the CLCuMuB cloned from infected and susceptible scions reveals that the variants of CLCuMuB cloned from the upper scion are 78 nucleotides shorter (deletion within the non-coding region) than the typical CLCuMuB isolates existing in lower infected scion. The alignment also discovered that the variants of CLCuMuB cloned from upper scion had an insertion of 27nt in the middle of βC1 ORF which could possibly increase the size of βC1 protein by 9 amino acids. The late induction of disease symptoms in upper susceptible scion suggests that only the variants of CLCuMuB with deletion of 78 nucleotides (coordinates 753–831nt) and an insertion of 27nt were able to move into G. arboreum. These variations in CLCuMuB may play an important role in the breaching of resistance and may have enabled the moment of betasatellite within G. arboreum. However, there is still need to carry out site directed mutagenesis study to find out the role of these 9 amino acids in resistance breaking. Begomoviruses can sustain host plant defenses by mutating their proteins that can be handy in maintaining viral cycle allowing them to bypass the host plant defenses as has been reported previously that mutation of a single amino acid in the replication associated protein of African cassava mosaic virus (ACMV) resulted in a differential host-mediated hypersensitive response to facilitate its replication (Jin et al., 2008).
Southern hybridization to detect the presence/titer of betasatellite in upper scion and G. arboreum revealed that the amount of betasatellite in the upper susceptible scion was significantly less than the level seen in the infected scions; however no such hybridization signals could be detected for the betasatellite in G. arboreum.
To our knowledge this is the first report of identifying and cloning ChLCuB from G. arboreum, a major breakthrough resulting in the breach of resistance in G. arboreum. The results indicated that the resistance of G. arboreum to CLCuD may be due to poor virus replication and interference in short distance movement. The information gained from this study may help in understanding the resistance mechanism of G. arboreum. The information gained from this study may be utilized in future breeding programs for the development of CLCuD resistant genotypes to alleviate the losses to cotton crops.
