Genetic Similarity between Cotton Leafroll Dwarf Virus and Chickpea Stunt Disease Associated Virus in India
Article information
Abstract
The cotton leafroll dwarf virus (CLRDV) is one of the most devastating pathogens of cotton. This malady, known as cotton blue disease, is widespread in South America where it causes huge crop losses. Recently the disease has been reported from India. We noticed occurrence of cotton blue disease and chickpea stunt disease in adjoining cotton and chickpea fields and got interested in knowing if these two viral diseases have some association. By genetic studies, we have shown here that CLRDV is very close to chickpea stunt disease associated virus (CpSDaV). We were successful in transmitting the CLRDV from cotton to chickpea. Our studies indicate that CpSDaV and CLRDV in India are possibly two different strains of the same virus. These findings would be helpful in managing these serious diseases by altering the cropping patterns.
Cotton leafroll dwarf virus (CLRDV) and chickpea stunt disease associated virus (CpSDaV) both are caused by Polerovirus (Family: Luteoviridae) and cause serious yield losses in the respective crops. Losses to the tune of 80% in susceptible varieties is reported from South America (Distéfano et al., 2010; Silva et al., 2008). Similarly, CpSDaV is the most serious virus problem in chickpea in India and in several other countries causing 80–95% yield loss (Kotasthane and Gupta, 1978; Reddy and Lava Kumar, 2004). Both the viruses are positive-sense, singlestranded RNA virus and are transmitted by aphids: Aphis gossypii in cotton (Corrêa et al., 2005), and Myzus persicae and A. craccivora in chickpea (Horn et al., 1996). Cotton plants affected by this disease show stunting, leaf rolling, intense green foliage, vein yellowing, brittleness of leaves, reduced flower and boll size, sometimes resulting in sterility of plants. Infected chickpea plants show leaf reddening, internode shortening, stunting and phloem browning in the collar region (Horn et al., 1996).
Occurrence of CLRDV in India was, for the first time reported by us (Mukherjee et al., 2012). Since both these viruses are caused by Poleoviruses and since there is an overlap in cotton and chickpea growing seasons in India, we investigated if both the infections (in chickpea and cotton) are caused by the same virus. Leaf samples from plants with symptoms suggestive of CLRDV infection in cotton and CpSDaV in chickpea, alongwith healthy looking plants were collected from cotton fields in Nagpur, Maharashtra, India and washed with RNase-free sterile double distilled water. RNA was extracted from leaf samples (100 mg) using Spectrum Plant Total RNA kit (Sigma, St. Louis, MO, USA). Reverse transcription PCR (RT-PCR) was performed using Qiagen OneStep RT-PCR kit (Qiagen, Foster City, CA, USA) following the manufacturers’ instructions using the primers PL4F (5′-GCGACAAATAGTTAATGAATACGGT-3′) and o3R (5′-GTCTACCTATTTBGGRTTNTGGAA-3′), as described earlier (Mukherjee et al., 2012). The primers amplify a region of approximately 600 bp of the capsid protein/movement protein sequence of CLRDV (Corrêa et al., 2005). PCR conditions were: denaturation at 94°C for 45 s, annealing at 49°C for 45 s, extension at 72°C for 45 s, for 35 cycles, and final extension at 72°C for 10 min (Mukherjee et al., 2012). The PCR products were sequenced directly and translated with EXPASY Translate tool (http://web.expasy.org/translate) and the sequences aligned on www.phylogeny.fr after curation (removal of poorly aligned regions) with GBLOCKS (http://phylogeny.lirmm.fr/phylo_cgi/one_task.cgi?task_type=gblocks). A phylogenetic tree was drawn vis-a-vis homologous sequences downloaded from the GenBank, using MEGA 5 software (Tamura et al., 2011). We also attempted transmission of the virus from infected cotton plants to chickpea plants. Aphids from infected cotton plants were released on healthy chickpea plants grown in pots in a net house under protected condition. The presence of the virus in these infected cotton (source of inoculum) and chickpea plants was confirmed by RT-PCR as described above, and the product sequenced and compared.
Clear symptoms of CLRDV and CpSDaV were seen simultaneously in respective fields growing side-by-side (Fig. 1). PCR from symptom-bearing samples resulted in the amplification of an approximately 600 bp fragment which is of the expected size (Fig. 2). PCR from healthy samples did not produce an amplicon. The CLRDV sequence has earlier been deposited by us in GenBank (accession No. JN033875). An alignment of the coat protein sequences from CLRDV and CpSDaV from this study and those reported earlier is presented in Table 1 and Fig. 3. The phylogenetic tree revealed genetic similarity among the Indian strains of CLRDV and CpSDaV, which were distinctly different from that of the Brazilian and Thailand strain of CLRDV (Fig. 4). Two distinct strains of CpSDaV associated with chickpea stunt disease was reported earlier at Hyderabad, India (Naidu et al., 1997). In the transmission experiment, chickpea plants infested with aphids from infected cotton plants incited typical symptoms in chickpea in pot experiments in net house (Fig. 5). The un-infested plants remained healthy. The capsid/movement protein sequence amplified by RT-PCR from these infected chickpea plants were very similar to that obtained from the cotton plants which served as source of inoculum. This experiment confirmed that the cotton strain can infect chickpea and incite similar symptoms as the chickpea strain. However, we could not transmit chickpea stunt virus to cotton plants (data not shown).
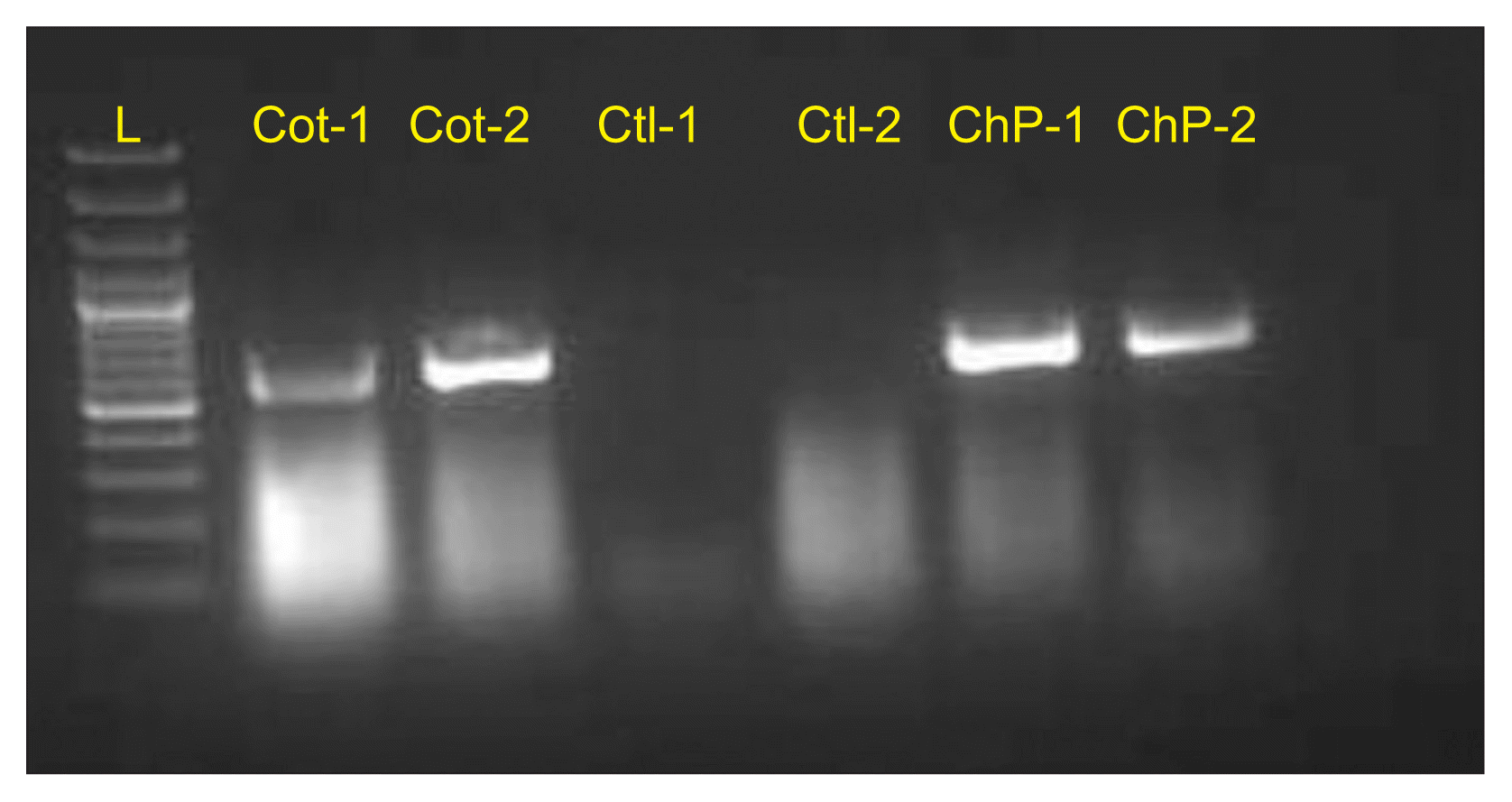
RT-PCR amplification of the coat/movement protein gene. Amplification, by RT-PCR, of coat/movement protein sequence of cotton leafroll dwarf virus (CLRDV) and chickpea stunt disease associated virus (CpSDaV) from infected cotton and chickpea plants collected from fields. L, DNA ladder; Cot, cotton; Ctl, healthy plants; ChP, chickpea.

Alignment of representative amino acid sequences corresponding to the partial coat protein gene of cotton leafroll dwarf virus (CLRDV) and chickpea stunt disease associated virus (CpSDaV). The alignment was performed after curing the sequences to remove the poorly aligned regions. Similar amino acids are shaded in gray.
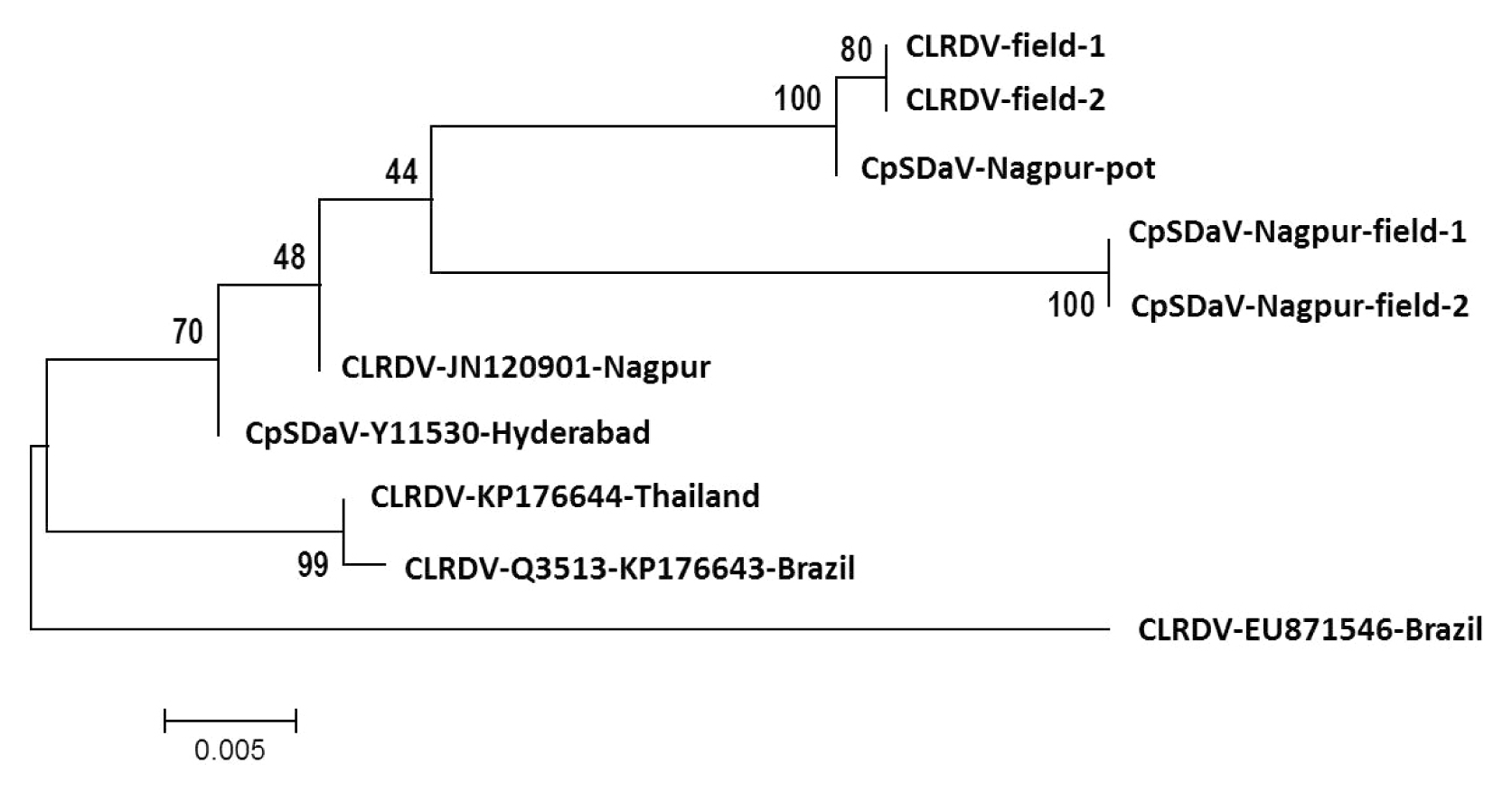
Phylogenetic tree showing the genetic similarities between partial coat protein sequences of cotton leafroll dwarf virus (CLRDV) and chickpea stunt disease associated virus (CpSDaV). The sequences were aligned by MUSCLE and curated by GBLOCKS on www.phylogeny.fr. Phylogeny reconstruction was done on MEGA 5 software (Tamura et al., 2011) by maximum likelyhood method with 1,000 bootstrap replications. The tree with the highest log likelihood (−681.1627) is shown. The tree, obtained by neighbor-joining method and BioNJ algorithms, is drawn to scale, with branch lengths measured in the number of substitutions per site.

Transmission of cotton leafroll dwarf virus from cotton to chickpea. Infestation of chickpea plants with aphids from infected cotton plants caused typical stunt symptoms in pot experiments in net house. Non-inoculated control is on the right.
Our findings suggest that leafroll dwarf virus from cotton can get transmitted to chickpea causing chickpea stunt. These findings have serious implications in Indian agriculture, since both the crops are grown simultaneously in vast areas. This calls for a modification of cropping patterns as a component of the disease management strategy.
Acknowledgments
Authors are thankful to TMC MM-1 for funding the work and Director, ICAR-Central Institute for Cotton Research, Nagpur for his support and encouragement.

