Characterization of a Begomovirus-Betasatellite Complex, Producing Defective Molecules in Spinach (Spinacia oleracea L.), a New Host for Begomovirus and Betasatellite Complex in Pakistan
Article information
Abstract
Spinach is a vegetable crop which is widely grown over a large area especially in Punjab province of Pakistan. Leaf curling and enations on spinach plant collected shown to be associated with the begomovirus Pedilanthus leaf curl virus (PeLCV) and Shahdadpur strain of Cotton leaf curl Multan betasatellite (CLCuMBSha). Defective molecules of half and quarter size derived from monopartite begomoviruses are usually generated by the deletion of virion-sense sequences. Characterization of defective molecules of PeLCV from spinach revealed that the molecules of half the size are derived from the deletion of complementary-sense genes while quarter size molecule appears to have evolved by further deletion. This is the first report of the begomovirus-betasatellite complex on spinach and unusual defective molecules derived from deletion of complementary-sense genes in Pakistan.
Geminiviridae is the largest family of insect-transmitted, single-stranded, circular plant DNA viruses, causing diseases to economically important food and fiber crops (Mansoor et al., 2003). The members of family Geminiviridae are divided into nine genera based on insect vector and genome organization of these viruses (Zerbini et al., 2017). Among them, whitefly-transmitted viruses, which are classified in the genus Begomovirus, constitute the largest and the most important group. Begomoviruses have emerged as a major constraint for food and fiber crops that are more common in tropical and sub-tropical climatic regions. The emergence of these plant viruses is attributed to the increase in vector population, expansion in host range, changes in cropping system (extensive agriculture) and exchange of germplasm and planting material. On the other hand, recombination among viral genomes contributes to the emergence of more virulent viruses which not only results in increased disease severity but also helps in acquiring new host for begomoviruses (Lefeuvre et al., 2007).
Begomoviruses consist of either two components named as DNA A and DNA B of 2.6–2.8 kb (bipartite) or are monopartite where a single genomic component equivalent of DNA A is sufficient to cause the disease. The vast majority of begomoviruses from the Old World (OW) consists of a single component associated with a satellite molecule referred to as betasatellite (Briddon and Stanley, 2006). Betasatellites are half of the size of the helper begomovirus (approximately 1.4 kb) encoding a single gene βC1 and depend on the helper virus for their replication, systemic movement, and vector transmission. These satellite molecules play an essential role in the induction of disease symptoms in the respective host plant. Betasatellites (previously known as DNA-β) do not share any significant sequence identity with their helper viruses except nonanucloetide sequence (TAATATTAC) found in all begomoviruses (Briddon et al., 2003). Begomovirus disease complexes caused massive losses to cash crops (Mansoor et al., 2006). However, the importance of weeds and other alternative hosts and their role in the spread of viruses in the absence of the main crop has been recognized lately (Jyothsna et al., 2011; Mubin et al., 2009; Mubin et al., 2010; Wyant et al., 2011).
In a kitchen garden, adjacent to a cotton field, severe leaf curl symptoms were observed on spinach near Central Cotton Research Institute (CCRI), Multan, Punjab, Pakistan in 2007. The infected plant was severely stunted with curling and crinkled leaves with enations like outgrowth. To identify the causative agent, total DNA was extracted from symptomatic leaves using CTAB method (Doyle and Doyle, 1990). A set of universal primers (BegomoF/BegomoR) that was designed to amplify begomoviruses was used in PCR for the amplification of begomovirus (Shahid et al., 2007). To confirm the association of betasatellite with the disease, universal primers (β01/β02) were used for the amplification of betasatellite through PCR (Briddon et al., 2002).
A PCR product of about 2.8 kb was amplified by universal primers for begomoviruses and confirmed the association of a begomovirus with the disease. However, besides specific full-length begomoviral PCR product, additional PCR products of approximately 1.4 kb and 0.7 kb were also amplified. With primers β01/β02 a product of ~1.4 kb was amplified. All amplified products were cloned into TA cloning vector pTZ57R/T (Thermo, USA) and were fully sequenced in both orientations for further analyses. Sequences were assembled and analyzed using the DNAStar program. The coding regions were identified using Open Reading Frame (ORF) Finder at NCBI (https://www.ncbi.nlm.nih.gov/orffinder/). Multiple sequence alignments were performed using ClustalW in MEGA6 software which was subsequently used to draw neighbor-joining dendrograms with 1000 bootstrap replicates (Tamura et al., 2013).
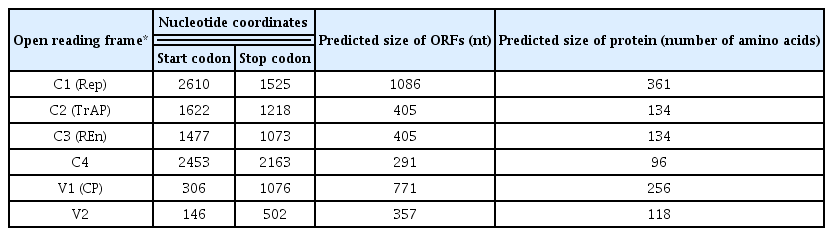
BLASTn analysis of complete genome sequence of the begomovirus (2757 bp) revealed 98% nucleotide sequence identity with Pedilanthus leaf curl virus (PeLCV) isolated from Glycine max ([Pakistan:Nawab Shah:06]-AM948961) confirming the begomovirus isolated from spinach as an isolate of PeLCV. The sequence was submitted to the nucleotide database database under the accession number HF568781. In concordance with other OW begomoviruses, the begomovirus isolated from spinach has all the open reading frames (ORFs) which are essential to perform necessary functions. The positions and predicted sizes of the encoded ORFs, two in the virion sense and four in complementary sense are presented in Table 1. Phylogenetic analysis of begomovirus based on the alignment of the complete nucleotide sequences of selected PeLCV sequences, which are available in the databases, is shown in Fig. 1. In phylogenetic dendrogram, the begomovirus isolated from spinach also showed a close relationship with the isolate of PeLCV isolated from Glycine max ([Pakistan:Nawab Shah:06]-AM948961).
Phylogenetic dendrogram based on the alignment of all Pedilanthus leaf curl virus (PeLCV) sequences. Vertical branches are arbitrary while the horizontal branches are proportional to calculated mutation distance. Values at nodes indicate percentage bootstrap values (1000 replicates). The isolate descriptors are given along with the host name. The tree was arbitrarily rooted on the sequence of Cotton leaf curl Gezira virus (CLCuGeV), which is an unrelated sequence of a similar size. The isolate described here is shown in bold.
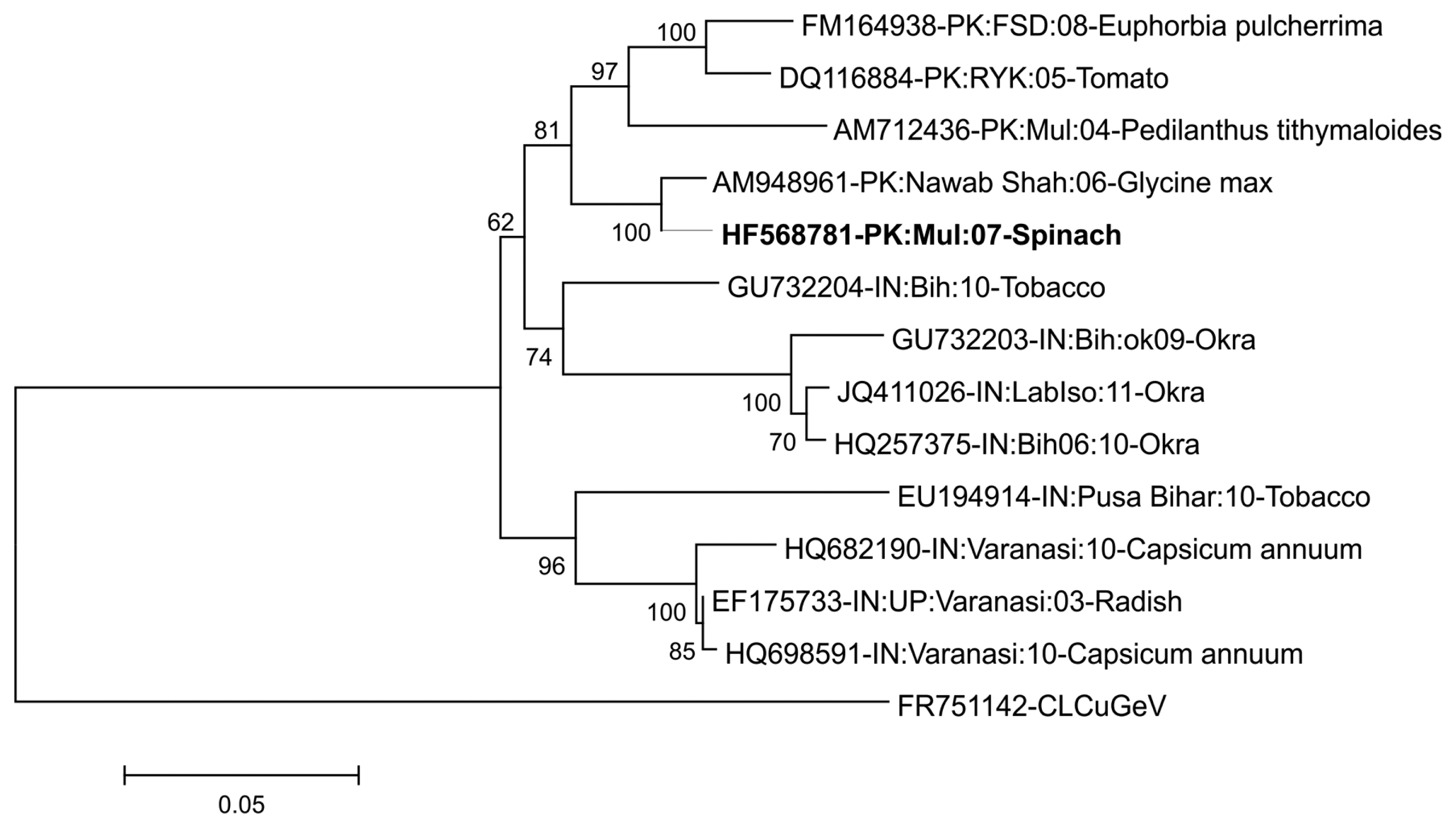
To confirm this, pairwise identity comparison of viral isolate from spinach was done by Sequence Demarcation Tool (SDT v1.2) (Muhire et al., 2014). Along with the studied begomoviral isolate, twelve sequences were obtained from the database, which were used for analysis. Based on the species demarcation criteria, which is 91% (Brown et al., 2015), these isolates were identified as PeLCV. According to SDT analysis, isolates from spinach showed maximum homology (98%) with the isolate from Glycine max (AM948961) from Nawab Shah, Pakistan (Fig. 2).
Nucleotide pairwise identity color matrix of PeLCV genomes. Complete description with the identity percentage of available PeLCV isolates in the database (calculated with Muscle algorithm) with the isolate described has been given with the representations of color. The isolate determined in this study is shown in the colored box. The unrelated sequence of CLCuGeV is also added to differentiate PeLCV isolates.
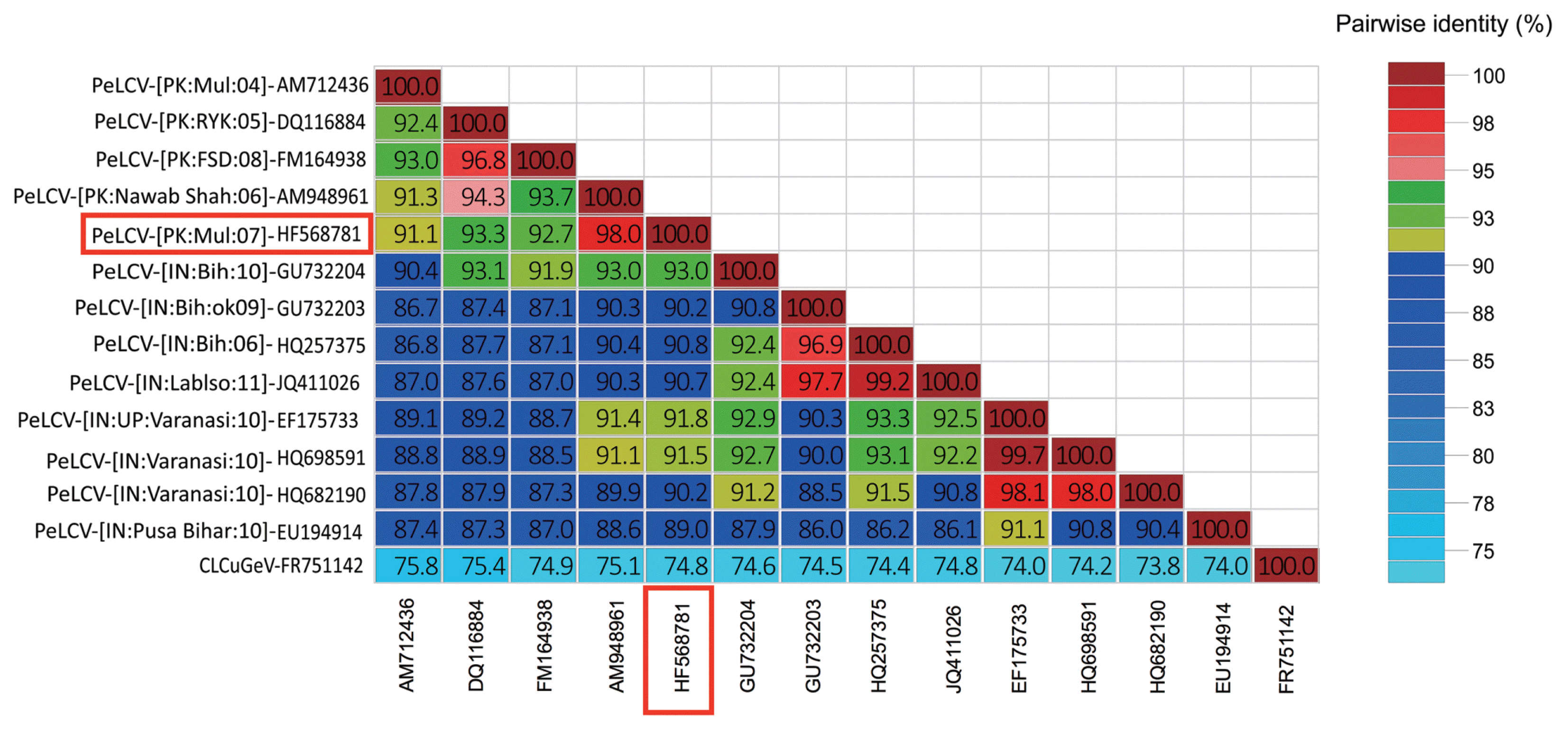
Defective (df) DNAs are often associated with the geminiviral diseases, which are derived from a partial deletion of viral genomes (Patil and Dasgupta, 2006). These defective molecules have nonanucloetide sequence, which suggests that these df molecules are trans-replicated by the helper virus, resulting in reduction of the viral symptoms, thus referred as defective interfering molecules (Patil and Dasgupta, 2006). Reduction in disease symptoms is either due to initiation of host RNAi mechanism (Bach and Jeske, 2014) or by the competition of df molecules with the helper virus for the cellular resources (Stanley et al., 1990). Along with the full-length begomovirus, two defective begomoviruses were also cloned from the sample. Complete sequencing of defective begomoviruses, having nonanucloetide sequence revealed DNA molecules of half and a quarter of the full-length begomovirus, 1459 and 714 nt respectively. The sequences are available in the database under accession numbers HF568782 and HF568783. Comparison of ORFs of df begomoviruses with full-length molecule provides information about deleted regions (Fig. 3). These df molecules have same conserved regions like V2, V1 (CP), and C1 (Rep). The size of V2 almost remains same in full-length and defective molecules although the other ORFs varies in length due to sizes of molecules (Table 2). Truncated Rep may initiate the replication of df molecules by a helper virus, thus helps in maintenance of df molecules. The role of the host plant is very important in the production of df molecules as it has been reported that experimental plants contained a significant amount of df geminiviruses when inoculated for long period of time which might be due to the high replication of the helper virus for long term infection (Liu et al., 1998). However, these df molecules restrict the virus from killing the host plant by inhibition of viral replication. It is noteworthy that these defective molecules were cloned during the efforts to identify and clone the full-length begomovirus from spinach plant under natural infection conditions. Sequence comparison of both df molecules showed that these are derived exclusively from their respective PeLCV (Fig. 4).
Diagrammatic representation of df begomoviruses in comparison with the full-length molecule. A full-length begomovirus prevalent in the region with all the genomic elements is shown at the top. Deleted regions in defective molecules are shown below the full-length molecule.

Features of df molecules isolated from spinach. Accession numbers and size of molecules is given in the top row of the table
Alignment of full-length PeLCV with both defective molecules analyzed through Clustal V using MegAlign-DNASTAR. Accession numbers of clones with their names is given on the left side while the position of nucleotides is given on right side. Nucleotide sequences of defective molecules that match with full-length PeLCV clone were shown in red highlighted color. The majority of consensus is shown at the top of sequences.
Betasatellites are a group of single stranded DNA satellites associated with most of the OW monopartite begomoviruses (Amin et al., 2006; Briddon and Stanley, 2006). By using universal primers, a product of 1347 bp was amplified through PCR which confirmed the association of betasatellite with the disease symptoms. Cloned product was sequenced and deposited in the database under accession number HF568784. Betasatellite sequence contains all the features of betasatellites; an ORF in the complementary sense orientation known as βC1, an adenine (A) rich region and a highly conserved region in all betasatellites known as satellite conserved region (SCR) containing the nonanucloetide sequence. βC1 is an important ORF that encodes a protein essential for pathogenicity, besides being important as a suppressor of gene silencing (Cui et al., 2005; Saeed et al., 2007).
The sequence comparison of the betasatellite isolated from spinach with available sequences in the database reveals that the isolated betasatellite has the highest sequence homology with previously isolated Cotton leaf curl Multan betasatellite (CLCuMB) isolate Dar-Beta-3 (Accession No. EU384603). βC1 gene of betasatellite isolated from spinach encodes a protein consists of 118 amino acids (coordinates 550–194) showing 99% identity with βC1 of CLCuMB. Based on the species demarcation criteria for betasatellites, which is 78% (Briddon et al., 2008), it is indicated that betasatellite isolated from spinach is an isolate of CLCuMB. Phylogenetic analysis based on the alignment of selected CLCuMB sequences obtained from the database is shown in Fig. 5. In phylogenetic analysis, the betasatellite isolated from spinach grouped with the CLCuMB-Shahdadpur strain (CLCuMBSha). The sequence comparison of SCR regions of CLCuMB to determine the recombinant fragment originating from Tomato leaf curl betasatellite (ToLCB)shows that the betasatellite characterized here containing 27 nt derived from ToLCB confirming the betasatellite as Shahdadpur strain of CLCuMB (CLCuMBSha) (Akhtar et al., 2014).
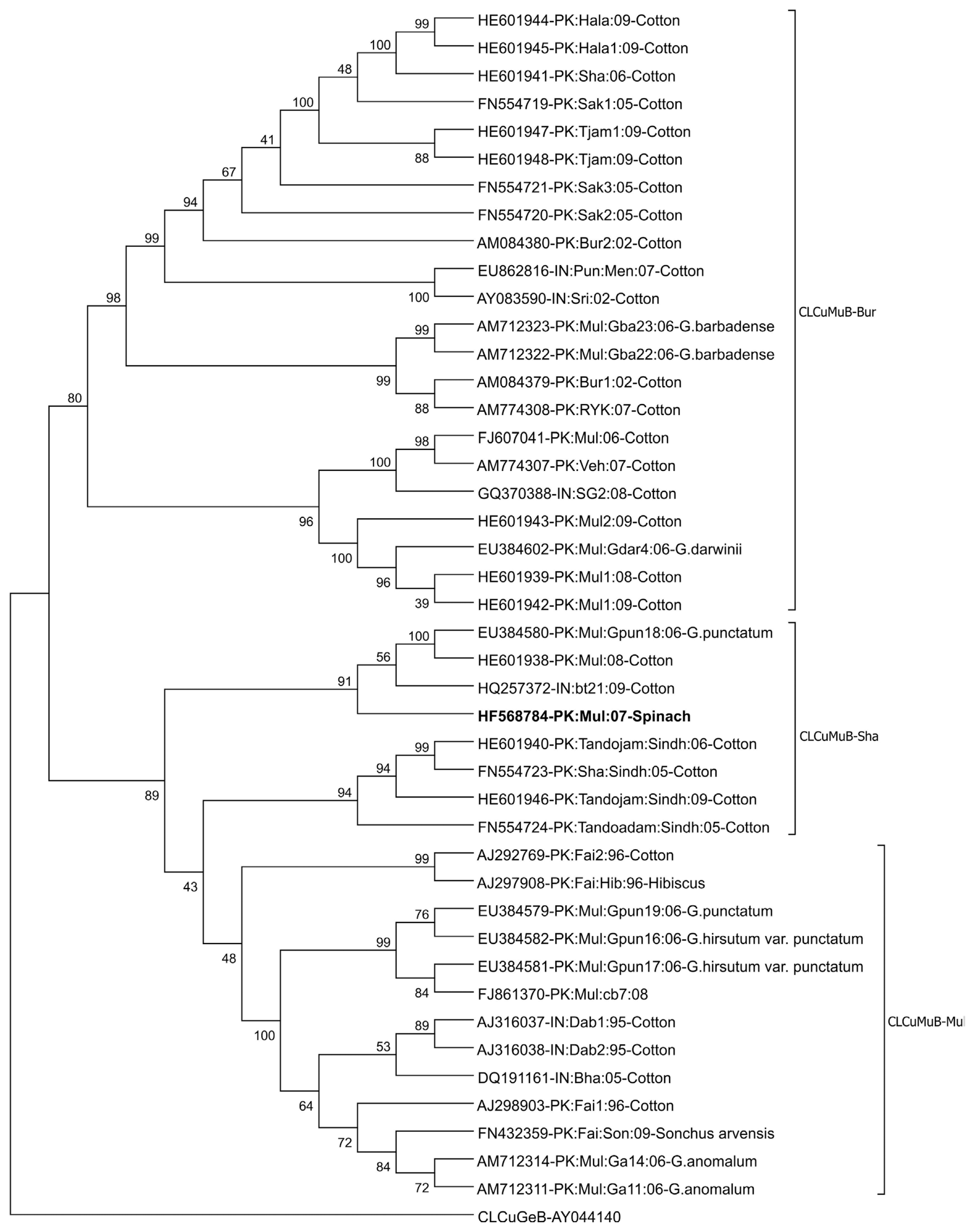
Phylogenetic dendrogram based on the alignment of selected betasatellite sequences. Vertical branches are arbitrary while the horizontal branches are proportional to calculated mutation distance. Values at nodes indicate percentage bootstrap values (1,000 replicates). The betasatellite acronyms used are Cotton leaf curl Multan betasatellite-Multan strain (CLCuMBMul), Cotton leaf curl Multan betasatellite-Burewala strain (CLCuMBBur) and Cotton leaf curl Multan betasatellite-Shahdadpur strain (CLCuMBSha). The tree was arbitrarily rooted on the sequence of Cotton leaf curl Gezira betasatellite (CLCuGeB), which is an unrelated sequence of a similar size. The database accession number along with the origin of sequences in each case is given. The isolate described here is shown in bold.
In begomovirus disease complexes, along with begomovirus and betasatellite, a self-replicating satellite-like component known as alphasatellite is also found to be associated. However, PCR amplification from spinach DNA extracts using universal set of primers for the alphasatellite of the most common monopartite disease complex present in Pakistan (CLCuD complex) was not successful. This indicates that the begomovirus infecting spinach is likely to be a monopartite begomovirus, lacking alphasatellite component.
The leaf curl infection symptoms found on spinach that belongs to the family Amaranthaceae are probably due to the high inoculum level of the virus in the adjacent cotton field as Multan is a major cotton growing area. However, few other vegetables are also grown in the field for local needs. Therefore, it is most likely that during cotton season whiteflies carrying the CLCuD complex are in high number and thus feeding on the other plant hosts as well that may result in the spread of this disease complex to other crops.
Beet curly top virus has been attributed as the cause of curly top disease in spinach showing the symptoms of yellowing, stunting, and curling (Baliji et al., 2004). Beet leafhopper is the vector responsible for the 1996 curly top disease epidemic of spinach in southwest Texas caused by Spinach curly top virus (SCTV) which was initially recognized as a Curtovirus species (family Geminiviridae) responsible for causing diseases in spinach (Baliji et al., 2007). However, Spinach severe curly top virus (SSCTV) is associated with the infected spinach in Arizona (Hernandez and Brown, 2010). Although recently another begomovirus Spinach yellow vein Sikar virus with the association of Spinach yellow vein betasatellite causing yellow vein disease in spinach has been reported from Rajasthan, India (Sahu et al., 2015). However, this is the first evidence showing the presence of PeLCV with cotton leaf curl disease associated betasatellite from diseased spinach plant from Pakistan. Further studies are needed to determine the impact of the disease. The identification of monopartite begomovirus with betasatellite from Indo-Pak region indicates that spinach is a host plant for begomovirus disease complex, although the extent of the disease is not known.
Acknowledgements
MNT was supported by a fellowship from Higher Education Commission (HEC), Government of Pakistan. We thank Dr. Rob W. Briddon for useful discussion.