Identification and Expression Analysis of Genes Induced in Response to Tomato chlorosis virus Infection in Tomato
Article information
Abstract
Tomato (Solanum lycopersicum) is one of the most widely grown and economically important vegetable crops in the world. Tomato chlorosis virus (ToCV) is one of the recently emerged viruses of tomato distributed worldwide. ToCV-tomato interaction was investigated at the molecular level for determining changes in the expression of tomato genes in response to ToCV infection in this study. A cDNA library enriched with genes induced in response to ToCV infection were constructed and 240 cDNAs were sequenced from this library. The macroarray analysis of 108 cDNAs revealed that the expression of 92 non-redundant tomato genes was induced by 1.5-fold or greater in response to ToCV infection. The majority of ToCV-induced genes identified in this study were associated with a variety of cellular functions including transcription, defense and defense signaling, metabolism, energy, transport facilitation, protein synthesis and fate and cellular biogenesis. Twenty ToCV-induced genes from different functional groups were selected and induction of 19 of these genes in response to ToCV infection was validated by RT-qPCR assay. Finally, the expression of 6 selected genes was analyzed in different stages of ToCV infection from 0 to 45 dpi. While the expression of three of these genes was only induced by ToCV infection, others were induced both by ToCV infection and wounding. The result showed that ToCV induced the basic defense response and activated the defense signaling in tomato plants at different stages of the infection. Functions of these defense related genes and their potential roles in disease development and resistance to ToCV are also discussed.
Tomato (Solanum lycopersicum L.) is one of the most widely grown and economically important vegetable crops in the world. Many pathogens cause disease in tomato and hinder production of tomato crops worldwide. Among these pathogens, viruses constitute one of the limiting factors for tomato production in many regions due to the absence of effective control measures. Recently, new viruses causing diseases in tomato has been reported and some of them are potentially threatening to tomato production in different regions of the world (Hanssen et al., 2010).
Tomato chlorosis virus (ToCV) is one of the recently emerged viruses infecting tomato, especially in greenhouses. Since it was first reported in Florida in 1998 (Wisler et al., 1998a), ToCV has been detected in many tomato production regions including the Mediterranean basin (Hanssen and Lapidot, 2012). ToCV is a member of the genus Crinivirus in the family Closteroviridae (Wisler et al., 1998b). It is a phloem-limited virus with a positive sense single-stranded bipartite RNA genome. The genome is encapsidated in flexuous virions of 800 to 850 nm in length and 12 nm in diameter (Wisler et al., 1998b). The RNA 1 genome is consisted of four different ORFs encoding six proteins associated primarily with viral replication. On the other hand, RNA 2 is organized into nine ORFs encoding HSP70 homolog, suppressor of gene silencing (p22) and two capsid proteins, the major coat protein (CP) and the minor coat protein (CPm). The CPm was implicated in vector transmission of ToCV (Dolja et al., 2006; Karasev, 2000; Wintermantel et al., 2005).
ToCV was originally detected in tomato and caused widespread epidemics and significant economic losses in tomato production. However, it was also detected in wild hosts Physalis ixocarpa, P. peruviana, S. nigrum and Datura stramonium (Lozano et al., 2004; Trenado et al., 2007; Vargas et al., 2011). More recent studies showed that the virus was able to naturally infect other Solanaceous crops such as sweet pepper (Capsicum annuum) (Barbosa et al., 2011; Lozano et al., 2004) and potato (S. tuberosum) (Fortes and Navas-Castillo, 2012; Freitas et al., 2012). Furthermore, the virus was also detected in other economically important crops such as lettuce (Orfanidou et al., 2014) and winter squash (Solórzano-Morales et al., 2011). The host range studies revealed that ToCV was able to infect plants from about 20 different families mainly including a number of other weed species (Orfanidou et al., 2014; Solórzano-Morales et al., 2011; Wintermantel and Wisler, 2006). These studies suggested while the virus is spreading in tomato field and greenhouses in different regions of the world, it is also adapting other wild and cultivated plants to expand the spectrum of its hosts.
A significant progress has been made for detection and molecular characterization of ToCV isolates from different countries (Dovas et al., 2002; Jacquemond et al., 2009; Kataya et al., 2008; Louro et al., 2000; Papayiannis et al., 2011; Trenado et al., 2007; Wisler et al., 1998b). However, studies on the pathogenesis, the disease development process and the virus-host interaction lagged behind largely due to the absence of a simple and efficient inoculation method. Since ToCV is not transmitted mechanically (Dovas et al., 2002; Wisler et al., 1998b), all experimental studies relay on whitefly transmission (Dalmon et al., 2009; Orfanidou et al., 2016; Wintermantel and Wisler 2006). Thus, studies involving vector transmission are generally labor intensive and time consuming due to difficulties associated with maintaining and handling of whitefly colonies. Therefore, alternative transmission and inoculation systems such as a inter-stock grafting were used for ToCV inoculation and testing for the resistance of cultivated and wild tomatoes and their hybrids to ToCV (García-Cano et al., 2010). Using this method, it was shown that S. lycopersicum containing commercial tomato cultivars is susceptible to ToCV inoculation, but one wild tomato and two hybrids were somewhat resistant to ToCV infection (García-Cano et al., 2010). However, the nature of the resistance was not determined and gene(s) involved in the resistance have not been identified yet. More recently, a simpler patch–grafting method was developed for experimental inoculation of tomato seedlings with ToCV and coupled with quantitative RT-PCR (RT-qPCR) assay for monitoring ToCV in inoculated tomato plants (Çevik et al., 2019). The results showed that ToCV was detected at 15 dpi and the virus titer increased until 40 dpi and decreased thereafter in graft-inoculated tomato seedlings. Thus, it was proposed as an efficient method for studying ToCV-tomato interaction under the controlled environments (Çevik et al., 2019).
Plant-virus interactions in tomato has been explored by transcriptome analysis using suppression subtractive hybridization (SSH) (Alfenas-Zerbini et al., 2009; Sahu et al., 2010), microarrays analysis (Catoni et al., 2009; Hanssen et al., 2011; Lucioli et al., 2016; Shahid et al., 2015) and RNA sequencing (RNA-seq) (Chen et al., 2013; Wang et al., 2015). While transcriptome analysis of tomato-virus interactions was largely focused on Tomato yellow leaf curl virus (TYLCV) and related DNA viruses (Chen et al., 2013; Lucioli et al., 2016; Sahu et al., 2010, Shahid et al., 2015; Wang et al., 2015), a few studies were conducted for identification and expression analysis of genes involved in tomato interaction with different RNA viruses at different scales (Alfenas-Zerbini et al., 2009; Catoni et al., 2009; Hanssen et al., 2011). The SSH was efficiently used for the identification of differentially expressed genes in response to virus infections in different plants (Liu et al., 2012; Pompe-Novak et al., 2005; Shi et al., 2005) including tomato (Alfenas-Zerbini et al., 2009; Sahu et al., 2010). A number of genes induced or repressed in response to virus infections were identified in tomato using SSH (Alfenas-Zerbini et al., 2009; Sahu et al., 2010).
In this study, patch-graft inoculation and SSH coupled with macroarray hybridization were used to explore ToCV-tomato interaction at transcriptome level. A cDNA library enriched with ToCV-induced genes was constructed from ToCV-inoculated and mock-inoculated leaves of tomato seedlings by SSH. Genes induced in response to ToCV infection in tomato were identified by macroarray hybridization and the changes in the expression of these genes were validated by RT-qPCR.
Materials and Methods
Plant materials
Seeds of Moneymaker tomato cultivar, widely used for maintaining ToCV isolates, was purchased from ZenGarden (İzmir, Turkey) and used as a host plant. Seeds were germinated in plastic vials containing peat, perlite and vermiculite mixture in a controlled plant growth chamber at 25ºC and 70% relative humidity and with 16/8 h photoperiod. The seedlings were transplanted into 25 × 10 cm pots containing the same mixture. They were grown in the controlled plant growth chamber at the same conditions until their stem diameters reached to about 0.5–1 cm.
ToCV Isolate
A ToCV isolate previously recovered from a greenhouse-grown tomato plant in Aksu District of Antalya province, AKSU8, (Akdura and Çevik, 2011; Yeşilyurt and Çevik, 2019) was used in this study. Moneymaker tomato cultivar was inoculated with the original isolate by inter-stock or patch grafting (Cevik et al., 2018; García-Cano et al., 2010). The isolate was maintained in Moneymaker tomato cultivar by periodic graft inoculations.
ToCV Inoculation
Phloem-tissue containing stem segments of about 1 cm in length were longitudinally excised from a ToCV-infected source tomato plant. At least 10 seed-grown Moneymaker tomato seedlings with a stem diameter of 0.5 cm were selected and their stems were longitudinally cut about 1 cm with a scalpel in two different places. The phloem-tissue containing stem segments from ToCV-infected tomato plants described above were place on similar cuts generated in the stems of healthy tomato seedlings. The grafts on stems of the healthy plants were wrapped with parafilm to maintain phloem contact for ToCV inoculation by grafting. Two grafts were performed in different parts of the stem of each tomato seedling. Another group of tomato seedlings were grafted with similar stem segments from a healthy tomato plant and used as control mock inoculation. All ToCV and mock-inoculated plants were tested for ToCV infection. Leaf samples were collected from ToCV or mock-inoculated plants at 30 days post inoculation (dpi), immediately frozen in liquid nitrogen and kept at −80°C.
RNA Isolations
Leaf samples were collected from ten ToCV or mock-inoculated tomato seedlings at 30 dpi. All ToCV and mock-inoculated plants were tested for ToCV infection. Then, leaf samples from all ToCV or mock-inoculated seedlings were bulked separately. Poly (A+) RNA and total RNA were isolated from ToCV or mock inoculated leaf samples using the FastTrack 2.0 mRNA Isolation Kit (Invitrogen, Carlsbad, CA, U.S.A.) and RNAeasy plant mini kit (Qiagen, Hilden, Germany), respectively, according to their manufacturer’s instructions.
Suppression Subtractive Hybridization
A SSH was performed by PCR-Select cDNA Subtraction Kit (TaKaRa, Kusatsu, Japan) using Poly (A+) RNA from ToCV or mock-inoculated control plants as previously outlined (Şahin-Çevik, 2013; Şahin-Çevik and Moore, 2006; Şahin-Çevik et al., 2017). For construction of the subtractive cDNA library containing genes induced by ToCV, a forward subtraction was applied using the tester cDNA from ToCV-inoculated leaf samples and the driver cDNA from the leaf samples of mock-inoculated control. All steps of SSH and library construction was performed according to PCR-Select cDNA Subtraction Kit manual (TaKaRa, Kusatsu, Japan), briefly described here. First, cDNA strand was synthesized from 2 μg Poly (A+) RNA using AMV reverse transcriptase and the second cDNA strand was synthesized to obtain double stranded cDNA. Then, cDNAs were fragmented into small pieces by RsaI restriction enzyme digestion and different adapter molecules were ligated to the both ends of digested cDNAs. And then, cDNAs from ToCV-inoculated samples were hybridized with excessive amount of cDNAs from mock-inoculated control samples twice to eliminate common cDNAs in both samples. At the end of hybridizations, unhybridized unique cDNAs in ToCV-inoculated samples were selected. Finally, two consecutive PCR amplifications were performed for normalization and amplification of the unique cDNAs from ToCV-inoculated samples.
Library construction
cDNA library was constructed by T-A cloning of the secondary PCR products into the pGEM-T easy cloning vector (Promega, Madison, WI, U.S.A.) using T4 DNA ligase (Promega, Madison, WI, U.S.A.) according to the manufacturer’s instructions. Then, Escherichia coli strain JM109 competent cells were transformed with ligations and grown on LB medium with 100 μg/ml ampicilin and 20 μg/ml X-gal and 50 μg/ml IPTG to obtain the subtracted cDNA library enriched with ToCV-induced genes. Bacterial colonies containing pGEM-T easy plasmids with inserts were randomly selected from the cDNA library. The colonies were stroke on a LB media supplemented as above and numbered to consecutive master plates containing each selected cDNA clones. All selected colonies were screened by a colony PCR using the universal M13 primers. Then, glycerol stocks were prepared for all clones containing pGEM-T easy plasmids with inserts and stored at −80°C freezer for future use.
Sequence analysis
Selection, sequencing and sequence analysis of clones from the subtractive library were conducted as previously described (Şahin-Çevik, 2013; Şahin-Çevik and Moore, 2006; Şahin-Çevik et al., 2017) and briefly outlined here. Bacterial colonies confirmed to contain pGEM-T easy plasmids with a tomato cDNA inserts were randomly selected from the subtracted cDNA library. Selected clones were sequenced one directionally by Sanger sequencing using the M13 forward universal primer. cDNA sequences obtained from selected clones were first trimmed at both ends and then vector sequences were cleaned. Sequences shorter than 50 bp were eliminated and excluded from further analysis. The remaining trimmed and cleaned cDNA sequences obtained from the cDNA library were compared with each other and contigs were established using Contig Express module of Vector NTI Advance 11.1 software (Invitrogen, Carlsbad, CA, U.S.A.). cDNAs containing identical sequences and/or forming contigs with each other were combined to eliminate redundancy. The nucleotide sequences of each cDNA were then compared with NCBI nucleotide and protein databases using BLASTN and BLASTX applications, respectively. The putative cellular functions of each cDNA sequence were determined based on the similarity scores obtained from both BLAST analyses.
Preparation of nylon cDNAs macroarrays
The expression of selected unique cDNAs were measured by macroarray hybridization using custom nylon membranes containing all selected cDNAs and appropriate controls. To prepare nylon macroarrays, constitutively expressed housekeeping genes including, β-actin, tubulin, glyceraldehyde phosphate dehydrogenase, ubiquitin, genes were selected as internal controls for normalization. Two human genes Glycogen 2 (GenBank accession number: NM_001184703) and troponin T type 1 (GenBank accession number: NM_003283) were selected as negative controls (Şahin-Çevik, 2013). All controls were amplified by two-step RT-PCR using PrimeScript RT-PCR kit (TaKaRa, Kusatsu, Japan) according to the manufacturer’s instructions using gene specific primers and appropriate annealing temperature for each primer set. Then, all selected cDNAs were amplified directly from liquid bacterial culture grown in LB medium inoculated from the glycerol stocks by PCR using NP1 (5′-TCGAGCGGCCGCCCGGGCAGGT-3′) and NP2 (5′-AGGGCGTGGTGCGGAGGGCGGT-3′) primers supplied in the PCR-Select cDNA Subtraction Kit (Clontech, Mountain View, CA, U.S.A.). To prepare nylon macroarrays, PCR amplified cDNAs of selected from the library and control genes were first denatured with 0.4 N NaOH. Then, equal amount of denatured PCR products of cDNAs, control genes and water sample were spotted in duplicates on Hybond N+ nylon membranes (Roche, Basel, Switzerland) using a Bio-Dot Microfiltration Apparatus (Bio-Rad, Hercules, CA, U.S.A.) by a vacuum application (Şahin-Çevik, 2013; Şahin-Çevik et al., 2017). Nylon macroarrays were prepared in duplicates for hybridization with probes from ToCV-inoculated and mock-inoculated samples.
Macroarray hybridization and expression analysis of cDNAs
The expression of cDNAs from ToCV-induced cDNA library was analyzed by macroarray hybridization with DIG labeled cDNA probes synthesized from RNA isolated from ToCV or mock-inoculated control plants as previously reported (Şahin-Çevik, 2013). First, DIG labeled cDNA probes were synthesized from Poly (A+) RNAs isolated from ToCV-inoculated and mock-inoculated control tomato seedlings by the Transcriptor reverse transcriptase (Roche, Basel, Switzerland) according to the manufacturer’s instructions. Briefly, DIG labeled cDNA synthesis was performed using 2 μg of poly (A+) RNAs, 50 μM oligo dT primer, DIG labeling mixture containing 1 mM dATP, dCTP, dGTP (each), 0.65 mM dTTP, and 0.35 mM DIG-11-dUTP, 20 U Protector RNase inhibitor and 10 U Transcriptor reverse transcriptase (Roche, Basel, Switzerland) at 50ºC for 60 min. Then, nylon macroarrays were prehybridized in a sealed bag at 68ºC for 30 min with agitation in a hybridization oven. An equal amount of DIG-labeled cDNA probes from ToCV-inoculated and mock-inoculated control plants were added into the mix and hybridized at 68ºC for 16 h as described above. The next day, nylon macroarrays were washed in 2× SSC with 0.1% SDS at the room temperature for 5 min with agitation. Then, a more stringent wash was performed in 0.1× SSC with 0.1% SDS at 68ºC for 15 min with agitation in the hybridization oven. After the washes, hybridization signals were detected by CSPD-Star substrate (Roche, Basel, Switzerland) supplied in DIG-chemiluminescent detection kit (Roche, Basel, Switzerland). Finally, the blots were wrapped and sealed in a plastic bag and signals were captured and analyzed by ChemiDoc-It chemiluminescent imaging system (UVP, Cambridge, England). Two biological replications were performed with probes synthesized from ToCV and mock-inoculated plants from two independent inoculation experiments.
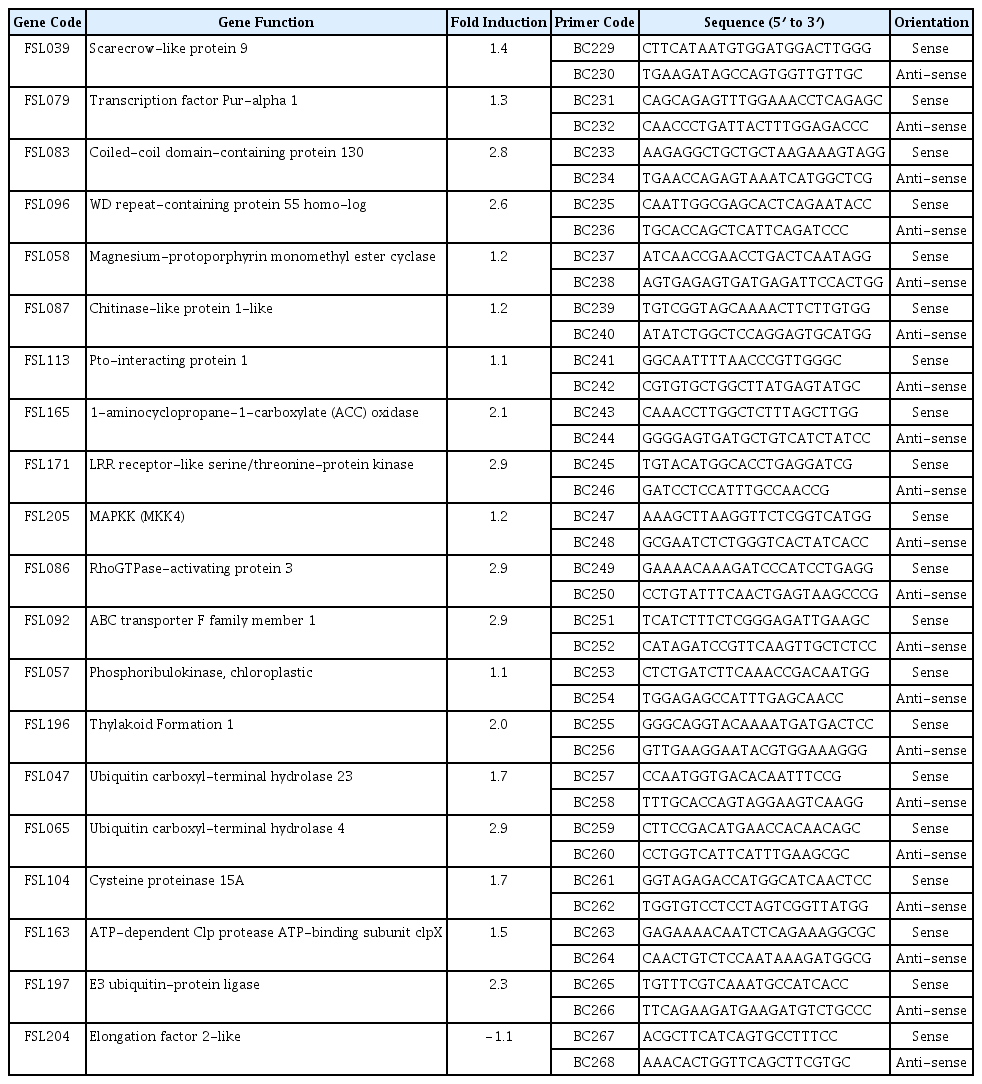
The gene expression data was obtained from hybridization of two duplicate nylon macroarrays with cDNAs and internal controls. One of the nylon macroarrays was hybridized with probes from ToCV-inoculated samples, the other one was hybridized with probes from mock-inoculated samples. The expression of individual cDNAs and controls in two replicates on the same membranes was determined after background subtraction by LabWorks software (UVP, Cambridge, England). The expression data obtained from multiple membranes from different experiments for each cDNA was normalized by the expression of reference genes used as constitutive controls on the same membrane. Then, the ratio of the normalized expression values of individual cDNA in macroarray hybridized with probes from ToCV-inoculated samples and probes from mock-inoculated samples was used as fold induction of the expression of individual cDNAs. Finally, log10 values of the normalized data were statistically tested for differences in the expression of each gene with ToCV-inoculated and mock-inoculated probes by a t-test (Şahin-Çevik and Moore 2006). cDNAs showing 1.5 or greater-fold increase in their expressions and with P ≤ 0.05 were considered to be induced in response to ToCV infection.
Validation of cDNA expression by real-time RT-PCR
Total RNA was isolated from leaf samples collected from ToCV-inoculated and mock-inoculated tomato seedlings at 30 dpi using RNAeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Then, 20 cDNAs showing increased expression in response to ToCV-inoculation in macroarray hybridization were selected. The expressions of twenty cDNAs were analyzed by two-step RT-qPCR using RevertAid cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, U.S.A.), SsoFast™ EvaGreen® Supermix (Bio-Rad, Hercules, CA, U.S.A.) and gene specific primers listed in Table 2. cDNAs were synthesized from 1 μg total RNA using by 10 units MMLV reverse transcriptase (Thermo Scientific, Waltham, MA, U.S.A.), 20 units RNase inhibitor and 50 μM oligo-dT primer. Selected cDNAs were amplified in 20 μl reaction mixture containing 1× SsoFast™ EvaGreen® Supermix (Bio-Rad, Hercules, CA, U.S.A.), 1 μl cDNA template and 10 μM of each gene specific primers. qPCR amplification of cDNAs was carried out along with β-actin as the reference gene. CFX96 Touch Real-Time System (Bio-Rad, Hercules, CA, U.S.A.) was programmed as 95ºC for 30 s for initial denaturation followed by 40 cycles of 95ºC for 10 s denaturation, 55–60ºC for 10 s primer binding and extension. The expression of each sample was determined at least two times in duplicates and expression was normalized using β-actin as the reference gene. Then, changes in the expression of ToCV-inoculated and mock-inoculated samples were calculated using previously described 2−ΔΔCt comparative CT method (Livak and Schmittgen, 2001) integrated into the CFX Manager™ software (Bio-Rad, Hercules, CA, U.S.A.).
Time course expression analysis of validated ToCV-induced cDNAs
The expression of 6 different ToCV-induced genes identified by macroarray hybridization and validated by RT-qPCR assay were analyzed in response to ToCV infection in detail. For this purpose, first, ToCV inoculation was performed by grafting to ToCV-sensitive Moneymaker tomato plants as described above. Leaf samples were collected from the inoculated and mock-inoculated control plants at 0, 1, 8, 15, 20, and 45 days post inoculation (dpi). Total RNA was isolated from all samples using RNAeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Then, cDNAs were synthesized from total RNAs using oligo (dT) primer and genes were amplified with gene specific primers (Table 2) using SYBR Green based RT-qPCR method as described above. Finally, the expression of 6 genes was determined at different stages of inoculation by 2−ΔΔCt comparative CT method (Livak and Schmittgen, 2001) using β-actin as the reference gene as described above.
Results
Identification of ToCV-induced genes from the subtracted cDNA library
A total of 240 cDNA clones were randomly selected from the subtracted cDNA library constructed using poly (A+) RNA isolated from both ToCV-inoculated and mock-inoculated tomato plants. Sequences of all selected cDNA clones were determined by unidirectional Sanger sequencing. No sequence was obtained from 18 of these clones and they were eliminated and excluded from further analysis. The length of cDNA sequences obtained from cDNA clones was ranged from 34 to 850 bp with an average of 569 bp. Similarity of sequences obtained from the subtractive cDNA library were compared with sequences in the GenBank databases using BLAST analyses. BLAST analyses showed that majority of the cDNA sequences showed homology with tomato genes available in the NCBI and Sol Genome databases. Some of the cDNA sequences showed homology with different parts of the same gene in the databases. Analyses also revealed that many cDNAs showed homology with previously characterized disease responsive genes from tomato and other plants. The sequences obtain from the library showed homology with genes involved in a various cellular functions and processes including transcription, defense, reponse and signaling, transport facilitation, energy, protein synthesis and fate and cellular biogenesis. The putative functions of ToCV-induced cDNAs from the library predicted by homologies determined by BLAST analyses were presented in Table 1. On the other hand, some of the cDNAs identified in the library were very similar or identical with sequences with unknown function and few others did not show any homology with any sequences in the databases; hence, they were grouped as cDNAs with unknown function.
Expression analysis of cDNAs by macroarray hybridization
A total of 108 cDNAs with non-redundant sequences were selected from the subtractive library for macroarray hybridization. Duplicate nylon macroarrays containing selected cDNAs and control genes were prepared and used to confirm the differential expression of these cDNAs in ToCV and mock inoculated tomato plants. The probes were synthesized from poly (A+) RNA isolated from pooled leaf samples of ten individual ToCV-inoculated or mock inoculated tomato plants. Duplicate macroarray membranes were hybridized with probes from ToCV or mock inoculated samples.
Macroarray hybridization showed that GYG2 and TNNT1 genes from human skeletal muscle and buffers used as negative controls did not show any signal in any of the membranes hybridized. However, housekeeping genes actin (ACT), tubulin (TUB), glyceraldehydes phosphate dehydrogenase (GADPH), ubiquitin (UBQ) used as internal control or reference genes were similarly expressed in both membranes hybridized with probes from ToCV-inoculated or mock-inoculated tomato plants indicating that the expression of the controls or reference genes was not affected by ToCV inoculation. On the other hand, the expression of cDNAs from the subtractive library was higher in macroarray hybridized with the probes from ToCV-inoculated plants than macroarrays hybridized with the probes from mock-inoculated control plants.
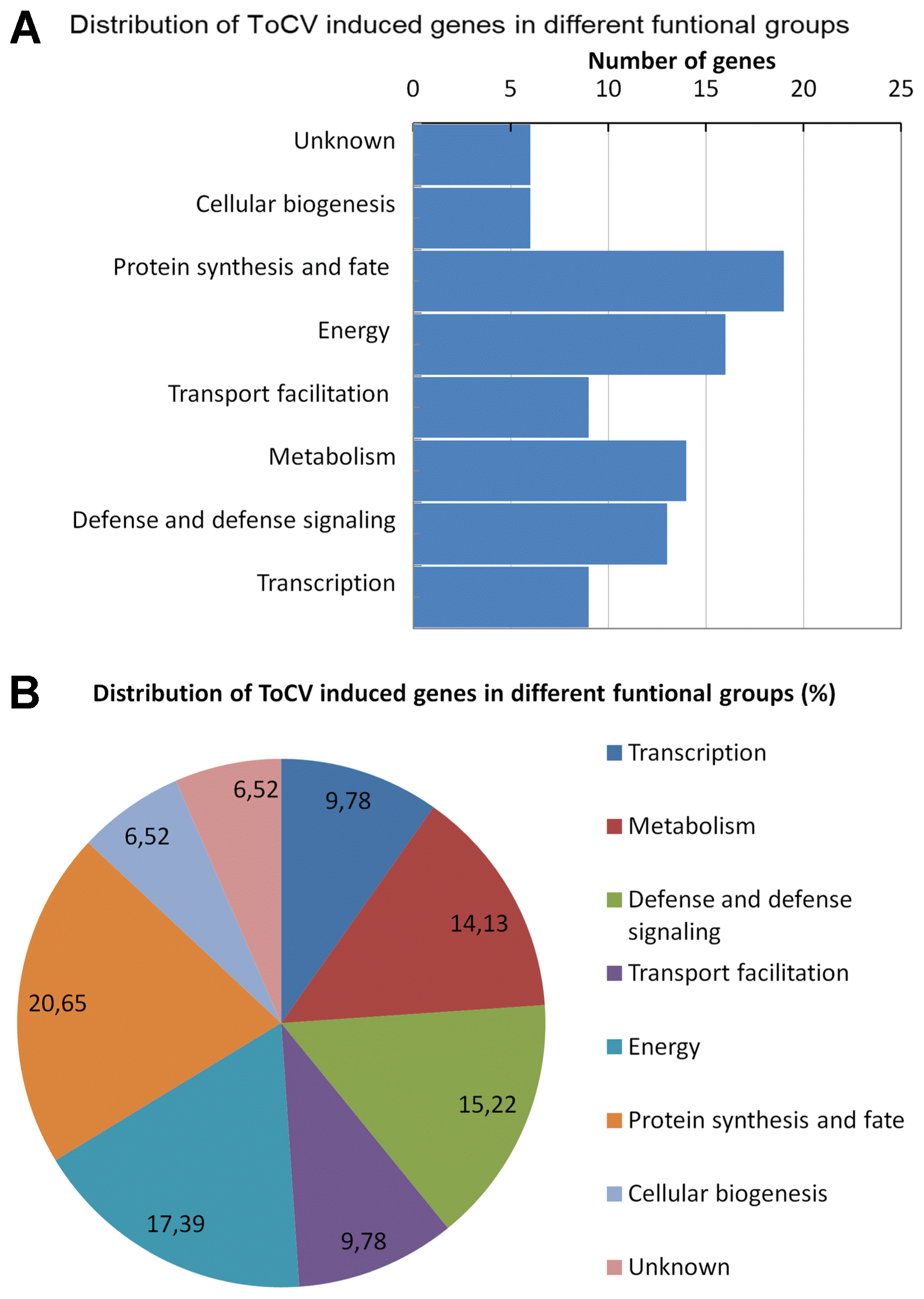
The expression of each cDNA was first quantified in macroarrays hybridized with ToCV-inoculated and mock-inoculated control. Based on the macroarray hybridization, a total of 99 cDNAs showed 1.5 to 3.58-fold increases in their expressions. All 99 ToCV-induced cDNA sequences were submitted to the GenBank under the accession numbers from JZ979249 to JZ979347. However, statistical test showed that induction of only 92 cDNAs with 1.5-fold or greater increase in their expressions were statistically significant at P ≤ 0.05. The expression analysis on macroarrays showed that more than 85% cDNAs selected from the subtractive library were significantly induced in response to ToCV infection. The ToCV-induced genes identified in this study were presented in the Table 1 with their Gen-Bank accession numbers, fold inductions, the putative functions and e-values.
ToCV-induced genes identified in this study were grouped into eight different functional groups based on the sequence identity or homology. The functions of ToCV-induced genes were related with following cellular processes; transcription (9); defense, and defense signaling (13); metabolism (14); transport facilitation (9); energy (16); protein synthesis and fate (19) and cellular biogenesis (6) (Table 1). The distribution of the ToCV-induced genes in different cellular process was shown graphically in Fig. 1. The results demonstrated that both subtracted cDNA library construction and macroarray expression analysis were functioned properly. The results also showed that a combination of the subtractive cDNA library construction and macroarray hybridization could be used as fast and efficient methods for identification of genes involved in biotic stresses in tomato and other plants.
Validation of gene expressions by real-time RT-PCR
The expression of some of ToCV-induced genes was analyzed by a RT-qPCR to confirm the differential expression in ToCV-inoculated and mock-inoculated control plants. A total of 20 cDNAs showing increased in their expressions in ToCV-inoculated tomato plants in macroarray analysis were selected from the subtractive cDNA library. The expression of 20 cDNAs listed in Table 2 was determined in ToCV-inoculated and mock-inoculated tomato plants by RT-qPCR using beta actin as the reference gene. The primers used for the expression analysis of selected cDNAs are presented in Table 2. RT-qPCR analysis showed that while the expression of β-actin used as the reference gene was not changed in ToCV-inoculated or mock-inoculated plants, the expression of selected cDNAs was affected by ToCV inoculation. The expression of 19 cDNAs were higher in ToCV-inoculated plants than mock-inoculated control plants (Fig. 2) and they showed 1.1–2.9-fold induction in response to ToCV inoculation. The highest level of increase was observed in the expression of cDNAs encoding Rho GTPase-activating protein 3 and ABC transporter family, probable leucine rich repeat (LRR) receptor-like serine/threonine protein kinase and ubiquitin carboxyl-terminal hydrolase 4 with 2.9-fold induction. On the other hand, with 1.1-fold induction phosphoribulokinase and Pto-interacting protein 1 showed the least increase in their expressions. The remaining genes were induced about 1.2 to 2.8-fold in response to ToCV inoculation (Table 2). However, the expression of Elongation factor 2 gene was reduced 1.1-fold in ToCV-inoculated plants. The expression results of macroarray hybridization were validated for 19 of 20 cDNAs by RT-qPCR assay confirming that the expression of these cDNAs were induced by ToCV inoculation at 30 dpi. Results of RT-qPCR assay were mostly in agreement with macroarray hybridization indicating that genes identified in this study are induced in response to ToCV-inoculation.
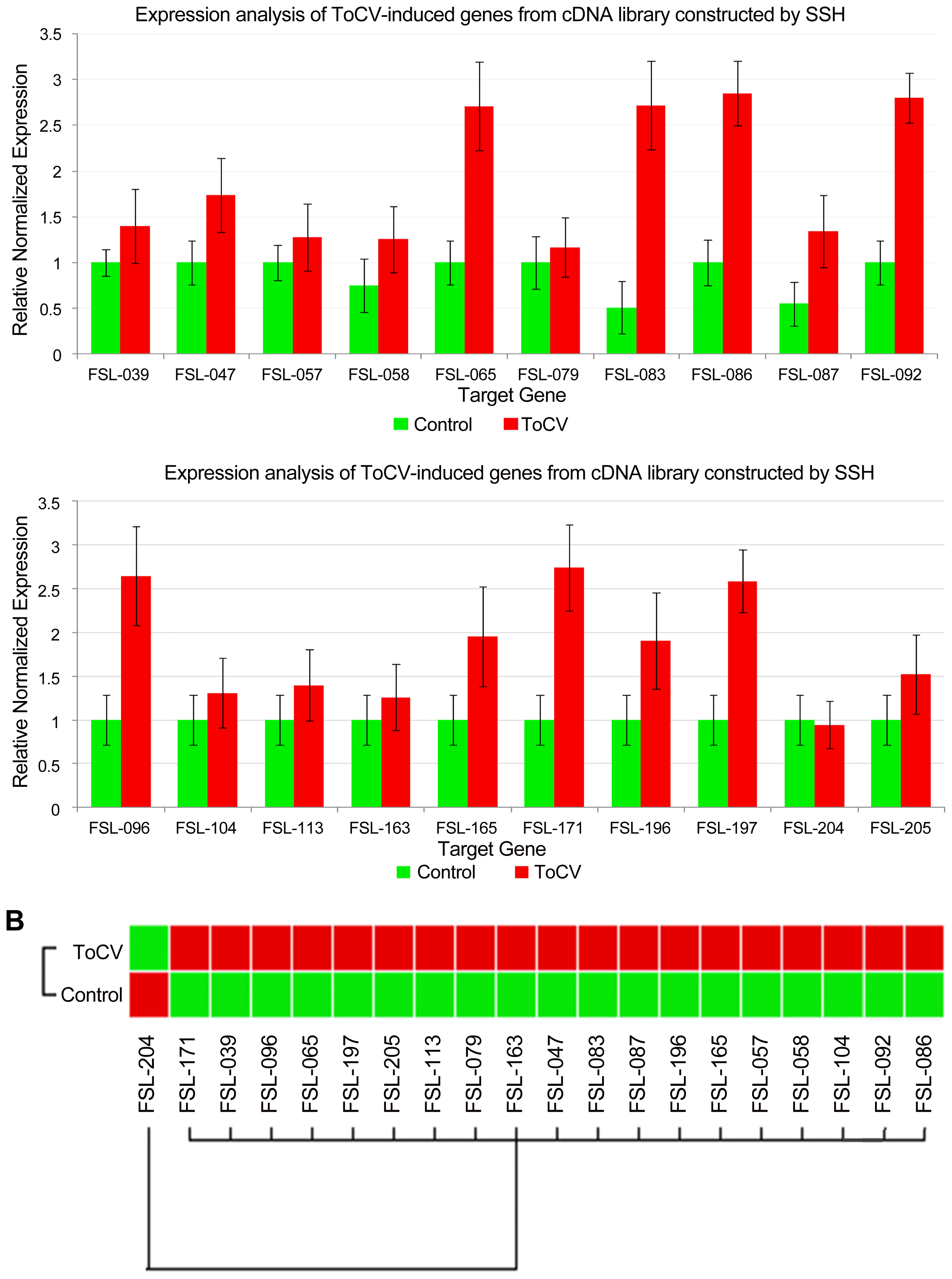
The expression analyses of 20 selected cDNAs induced in response ToCV infection in tomato by RT-qPCR. (A) The expression of each gene was determined and normalized with β-actin used as the reference gene in 30 dpi in inoculated (ToCV) and mock inoculated (Control) plants. Changes in the expression of each gene are shown as normalized fold induction in the graph. (B) Heat map of the expression of selected cDNAs in response ToCV-inoculation (ToCV) and mock-inoculation (Control).
Time course expression analysis of some ToCV-induced genes
Expressions of six ToCV-induced genes including scarecrow-like protein 9, WD repeat-containing protein 55, Rho GTPase-activating protein 3, probable LRR receptor-like serine/threonine-protein kinase, ubiquitin carboxyl-terminal hydrolase 4, elongation factor 2 were studied in response to ToCV infection in inoculated and mock-inoculated control plants at different dpi. The expressions of Sl-FSL 065 ubiquitin carboxyl-terminal hydrolase, Sl-FSL 171 Rho GTPase-activating protein 3-like and Sl-FSL 096 WD repeat-containing protein 55 homolog were not induced in mock inoculated plants, but their expressions were induced from 2 to 6-fold at 1 to 15 dpi in ToCV-inoculated plants (Fig. 3) suggesting that they are virus-induced genes. However, the expression of Sl-FSL 086 probable LRR receptor-like serine/threonine-protein kinase and Sl-FSL039 scarecrow-like protein was induced in both mock and ToCV-inoculated plants indicating that they are wound responsive genes. On the other hand, while the expression of Sl-FSL204 elongation factor 2-like gene was induced 2- to 3-fold at 15 and 20 dpi, respectively in mock-inoculated plants in response to wounding, its expression was induced slightly at 20 and 45 dpi in the ToCV-inoculated plants suggesting that this gene was induced by both wound and virus infection. The results of the time course expression analysis showed that while the expression of three genes was induced by only ToCV-infection, the other three genes were likely induced both by wounding and ToCV inoculation at different time points.
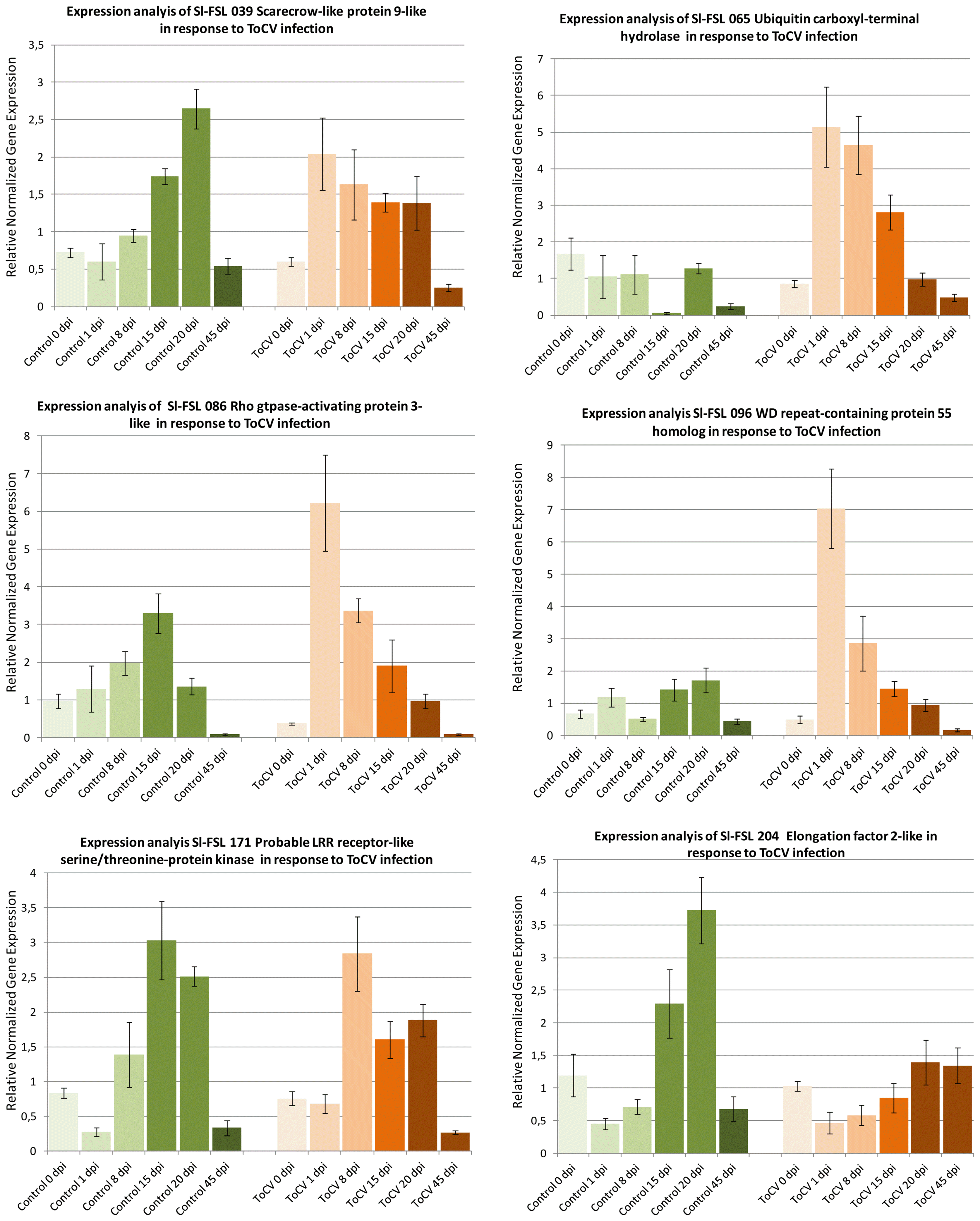
Time course expression analyses of 6 selected cDNAs induced in response ToCV in tomato by RT-qPCR. The expression of each gene was determined at different time points as 0, 1, 8, 15, 20 and 45 dpi and normalized with β-actin used as the reference gene in inoculated (ToCV) and mock inoculated (Control) plants. Changes in the expression of each gene are shown as normalized fold induction in the graph.
Discussion
Transcriptome analysis has become a very useful approach for studying the molecular mechanisms involved in plant-virus interaction in tomato and other plants. Different transcriptomics tools including SSH (Alfenas-Zerbini et al., 2009; Sahu et al., 2010), microarrays analysis (Catoni et al., 2009; Lucioli et al., 2016; Shahid et al., 2015) and RNA sequencing (RNA-seq) (Chen et al., 2013; Wang et al., 2015) have been used for identification and expression analysis of genes involved in tomato interaction with different viruses at different scales. The SSH has been proven as a useful tool for identification of differentially expressed genes in response to virus infection in citrus (Liu et al., 2012), maize (Shi et al, 205) potato (Pompe-Novak et al 2005) and tomato (Alfenas-Zerbini et al., 2009; Sahu et al., 2010). SSH has provided genome wide information on tomato-virus interaction by determining the changes in gene expression in response to virus infection and identification of genes involved in the plant defense pathways activated by virus infection in tomato (Alfenas-Zerbini et al., 2009; Sahu et al., 2010).
In this study, the SSH approach was used to explore ToCV-tomato interaction, and cDNA library enriched with ToCV-induced genes was constructed. A total of 265 ESTs were sequenced from the cDNA library constructed from the leaves of ToCV- and mock-inoculated tomato plants. Macroarray hybridization analysis resulted in identification of 92 non-redundant ToCV-induced cDNAs indicating that SSH coupled with reverse northern blot analysis functioned properly and it is an effective approach for identification of ToCV-induced genes in tomato as well as other biotic stress responsive genes in other plants. Based on the predicted or putative function, ToCV-induced genes identified in this study were divided into eight different groups including transcription (9.78%), defense response and defense signaling (14.13%), metabolism (15.21%), transport facilitation (9.78%), energy (17,49%), protein synthesis and fate (20.65%) and cellular biogenesis (6.52%). Induction of genes related with similar cellular processes were also reported in response to DNA (Miozzi et al., 2014; Sade et al., 2015) and RNA (Catoni et al., 2009; Hanssen et al., 2011) viruses in tomato as well as in response to the other viruses in other plants including potato (Pompe-Novak et al., 2005), Arabidopsis (Whitham et al., 2003) and tobacco (Lu et al., 2012). The number and functional grouping of genes identified in this study indicated that ToCV causes similar impact on tomato plants by re-organizing the host transcription, protein synthesis and metabolisms for increasing energy production to accommodate virus replication.
The induction of defense response is one of the main consequences of virus infection in plants. Plant viruses trigger different defense response in their host by inducing the expression of genes involved in hypersensitive reaction (HR), or systemic acquired resistance (SAR) based on the interaction with their host plant. Functional categorization of ToCV-induced genes showed that expressions of 13 different genes related to plant defense and/or defense signaling were induced in response to ToCV infection. Defense related genes identified in this study were associated with HR, ethylene biosynthesis, some component of SAR and defense signaling.
The HR initiated by incompatible interaction of pathogen avr gene(s) and R genes in the host plants. Since Moneymaker tomato cultivar used in this study does not contain a specific R gene against ToCV, genes involved nitric oxide (NO) and reactive oxygen species (ROS) production triggering HR response were not identified in this study suggesting that ToCV-tomato has a compatible interaction with tomato and does not induce the HR response as expected. The expression of a temperature-induced lipocalin (TIL)-like gene was induced in response to ToCV in this study. It was reported that a chloroplast lipocalin, the AtCHL protein prevents lipid peroxidation and protects Arabidopsis plants against oxidative stress (Levesque-Tremblay et al., 2009). Furthermore, the expression of developmentally regulated lipocalin-like gene, SlVRSL, was increased in response to TYCLV infection in resistant tomato plants. In addition, silencing of SlVRSL, lipocalin-like gene abolished TYLCV resistance and induce a necrotic response along the stem and petioles accompanied by the ROS production in tomato in response TYLCV infection. Unlike previously identified tomato lipocalin genes, SlVRSL was only induced in response to virus infection, but it was not responsive to neither cold, heat or salt (Sade et al., 2012). Increased expression of a TIL-like gene in response to ToCV in this study indicates that the TIL-like gene is a pathogen responsive and might have a role in suppression of ROS and HR in tomato. Similarly, expression of two rhodandase/sulfur transferase proteins, rhodanese-like domain-containing protein 14 and thiosulfate sulfur transferase 16 was induced in response to ToCV infection in this study. Rhodanese has thiosulfate sulfurtransferase domain, involved in sulfur metabolism in various organisms including plants (Cipollone et al., 2007). While these proteins are mainly play a role in elimination of toxic cyanogenic compounds in mammalian cells, the rhodandase/sulfurtransferase proteins also involved in detoxification of ROS in plants (Papenbrock et al., 2011). In addition, a host protein interacting both with N resistance protein from tobacco and p50 effector protein from TMV, named N-receptor-interacting protein 1 (NRIP1), was identified from tobacco plant. And the NRIP1 protein has a rhodandase/sulfurtransferase domain and activity and it was required for N-mediated resistance to TMV (Caplan, et al., 2008). This indicated that rhodandase/sulfur transferase proteins play an important role in plant defense response to virus infection. Induction of the expression of a two rhodandases in response to ToCV infection implies that these proteins may participate in recognition of pathogens or effectors or elimination of ROS.
Chitinases are produced as pathogenesis-related (PR) proteins in plants in response to infection with pathogens or exposure to the elicitors or phytohormone such as ethylene and salicylic acid (Hamid et al., 2013). As in other plants, chitinase are generally induced in response to infection with fungal pathogens in tomato and a chitinase specifically induced in response to Fusarium oxysporum f. sp. lycopersici was identified in tomato (Amaral et al., 2012). In addition, it was previously reported that chitinases were also induced in response to viral infection in tobacco (Bol et al., 1990). Identification of chitinase induced in response to ToCV infection in tomato suggest that chitinases may be induced directly or through SA-dependent pathway in response to virus infection in tomato. The expression of chitinase genes in response to tomato should be further investigated to elucidate the role of chitinase in virus infection in tomato and other plants.
Ethylene is a plant produced hormone playing roles in various biological processes, including, wounding and pathogen infection (Abeles et al., 1992; Zanetti et al., 2002). The 1-amino cyclopropane-1-carboxylate (ACC) oxidase catalyzes the final step of ethylene biosynthesis and is the key regulatory enzymes ethylene biosynthesis pathway. Several studies showed that the expression of the ACC oxidase gene was induced in response to infection with various pathogens in tomato (Van de Poel and Van Der Straeten, 2014). In this study, the ACC oxidase was induced in response to ToCV infection indicating that it may have roles in ToCV infection in tomato. In addition, acireductone dioxygenase (ARD) gene involved in methionine salvage pathway which recycle methionine for ethylene biosynthesis, was also induced in response to ToCV infection in tomato in this study. The ARD converts acireductone to provide substrate for the ACC oxidase and subsequently for S-adenosylmethionine (AdoMet) which is formed as a side-product of the ACC synthase in the ethylene biosynthetic pathway (Bürstenbinder et al., 2007). An ARD gene, OsARD1 from rice was identified and its involvement in recycling of the ethylene precursor AdoMet was shown in rice. The expression analysis of the OsARD1 gene also showed that it was an immediate-early ethylene responsive gene which plays a role in early feedback activation of the methionine cycle to maintain the ethylene synthesis with low levels of ethylene (Sauter et al., 2005). Induction of both the ACC synthase and the ARD1 genes in response to ToCV showed also that virus infection induced key genes in ethylene biosynthesis and maintenance in tomato indicates that ToCV infected plants produce and try to maintain high level of ethylene. However, direct analysis of ethylene in the infected plants is required to validate the results obtained from the gene expression studies.
A BAG family molecular chaperone regulator 6 (BAG6) was identified among ToCV-induced genes in this study. The BAG family molecular chaperone regulator involved in the modulation of different cellular processes including programmed cell death and stress responses (Kang et al., 2006). This family of proteins contains conserved BAG domain that interacts with molecular chaperones (Hsp70 and Hsc70) to regulate their activities (Doukhanina et al., 2006). More recently, the involvement of the BAG6 in basal resistance and autophagy was demonstrated in Arabidopsis (Li et al., 2016). It was shown that cleavage of the BAG 6 by an aspartyl protease is necessary for autophagy and plant defense against a fungal pathogen (Li et al., 2016). The BAG 6 of tomato was induced at 1 dpi then gradually decreased in response to ToCV infection in tomato in this study. Although the involvement of the BAG proteins to a fungal plant pathogen was reported (Li et al., 2016), the BAG6 is the first BAG protein induced in response to virus infection in plants. It was previously reported host hsp70 chaperons were recruited by some animal and plant viruses (Alam and Rochon, 2016). Since ToCV encodes its own hsp70 proteins, whether the BAG6 was induced by ToCV in tomato recruits and binds to viral or host hsp70 protein requires further studies.
Various proteins were involved in signaling in plant defense at different stages in response to different pathogens of effectors. Receptor-like protein kinases (RLK) gene-family contains variable cell surface and cytoplasmic receptors involved in signal reception. Three different RLKs including a probable inactive receptor kinase, a serine/threonine-protein kinase ATM, a probable LRR receptor-like serine/threonine-protein kinase genes were also induced in response to ToCV infection in this study. The RLKs interact with a diverse group of proteins playing a central role in signaling upon pathogen recognition and the subsequent activation of plant defense response (Afzal et al., 2008). RLKs play crucial roles in plant growth and development and immune system. Plants encodes large number RLKs as pattern recognition receptors (PRRs) that detect pathogen and/or host-derived molecular patterns as the first layer of inducible defense response in plants (Greeff et al., 2012). The RLK families are divided into sub-classes according to the structure of their N-terminal extracellular domain defining their ligand specificity. A LRR domain containing LRKs are the most common and well-characterized group (Tang et al., 2017). Plant RLKs regulate plant immunity by PRRs and initiate PAMP-triggered immunity (PTI) consisting of reactive oxygen species (ROS) production, callose deposition, generation of secondary messengers and induction of the expression of defense related genes. In addition, the RLKs are also involved in activation of several mitogen-activated protein (MAP) kinases (Greeff et al., 2012). The RLKs directly link diverse PPRs of bacterial and fungal pathogens to activate the MAP kinase cascades in Arabidopsis (Bi et al., 2018). The induction of different RLKs and downstream MAP kinases in response to ToCV infection in tomato suggests that some components of PTI were activated by ToCV infection in tomato.
The Rho family of small GTPases serve as the regulator of signal transduction in diverse cellular functions ranging from growth and development to adaptation to various environmental stresses in animals and plants (Kawano et al., 2014; Moon and Zheng, 2003). Recent evidence indicated that the Rho GTPase family plays a critical role in plant immunity (Kawano and Shimamoto, 2013; Kawano et al., 2014). The roles of two small Rho GTPases in rice (Oryza sativa, OsRac1), and barley (Hordeum vulgare, HvRacB) were elucidated (Kawano et al., 2014). They bind to cytoplasmic domains of RLKs and activate downstream defense response in both plants. ToCV induction of the expression of Rho GTPase-activating protein 3 indicated that some component of tomato defense system was activated in response to ToCV infection in tomato.
MAPK are the other key regulatory proteins mediating transduction of extracellular signals into intracellular components. They are involved in various signal transduction pathways and play key roles in plant defense against pathogen infection (Afzal et al., 2008). While many MAPKs were identified in tomato genome, the functions of these proteins have not been determined yet (Kong et al., 2012). The expression of SlMPK4 and SlMKK 4 was induced in response to Botrytis cinerea infection and also by exogenous application of jasmonic acid and ethylene precursor 1-amino cyclopropane-1-carboxylic acid (Li et al., 2014; Virk et al., 2012). The MKK4 gene was also identified in this study and the level of its expression was increased in response to ToCV infection indicates that MKK4 gene may have a role in response fungal pathogen as well as virus infection in tomato.
ToCV-tomato interactions were analyzed in the transcriptome level at the first time in this study. Although only the portion of the transcriptome was analyzed by SSH, 92 tomato genes were significantly induced upon infection with ToCV in tomato. The expression of some ToCV-induced genes was confirmed with different methods and at various time points of ToCV infection. The majority of the genes identified in this study were involved in maintaining basic cellular process such as protein synthesis, biogenesis, metabolism, energy production and transport in the cell. More than 10 genes identified in this study were previously reported to be involved in plant defense response against different pathogens suggesting that general defense pathways were activated by ToCV infection in tomato. While induction of a few of these genes were previously reported in response to virus infection, majority of defense related genes identified in this study was reported previously to be induced by fungal and bacterial pathogens in different plants. Therefore, expression of individual ToCV-induced genes identified in this study should be further analyzed in response to infection with ToCV and other viral, bacterial and fungal pathogens in tomato.
Acknowledgments
This study was conducted by a grant from the Scientific and Technological Research Council of Turkey (TÜBİTAK) project number 111O646.