siRNAs Derived from Cymbidium Mosaic Virus and Odontoglossum Ringspot Virus Down-modulated the Expression Levels of Endogenous Genes in Phalaenopsis equestris
Article information
Abstract
Interplay between Cymbidium mosaic virus (CymMV)/Odontoglossum ringspot virus (ORSV) and its host plant Phalaenopsis equestris remain largely unknown, which led to deficiency of effective measures to control disease of P. equestris caused by infecting viruses. In this study, for the first time, we characterized viral small interfering RNAs (vsiRNAs) profiles in P. equestris co-infected with CymMV and ORSV through small RNA sequencing technology. CymMV and ORSV small interfering RNAs (siRNAs) demonstrated several general and specific/new characteristics. vsiRNAs, with A/U bias at the first nucleotide, were predominantly 21-nt long and they were derived predominantly (90%) from viral positive-strand RNA. 21-nt siRNA duplexes with 0-nt overhangs were the most abundant 21-nt duplexes, followed by 2-nt overhangs and then 1-nt overhangs 21-nt duplexes in infected P. equestris. Continuous but heterogeneous distribution and secondary structures prediction implied that vsiRNAs originate predominantly by direct Dicer-like enzymes cleavage of imperfect duplexes in the most folded regions of the positive strand of both viruses RNA molecular. Furthermore, we totally predicted 54 target genes by vsiRNAs with psRNATarget server, including disease/stress response–related genes, RNA interference core components, cytoskeleton-related genes, photosynthesis or energy supply related genes. Gene Ontology classification showed that a majority of the predicted targets were related to cellular components and cellular processes and performed a certain function. All target genes were down-regulated with different degree by vsiRNAs as shown by real-time reverse transcription polymerase chain reaction. Taken together, CymMV and ORSV siRNAs played important roles in interplay with P. equestris by down modulating the expression levels of endogenous genes in host plant.
Phalaenopsis equestris, a butterfly orchid, are an important part of the global floriculture trade in cut or potted flowers. Cymbidium mosaic virus (CymMV; genus Potexvirus) and Odontoglossum ringspot virus (ORSV; genus Tobamovirus) severe affect the quality or value of P. equestris (Gibbs, 2000; Koh et al., 2014; Zettler et al., 1990). P. equestris simultaneously infected with both viruses normally showed severe symptoms, such as chlorotic, ringspots, and necrotic on plant leaves (Wong et al., 1994). Although much progress has been made about these two pathogen systems, interaction between P. equestris and viruses remains largely unknown, which led to deficiency of effective measures to control disease of P. equestris caused by infecting viruses.
Recently, next-generation high-throughput parallel sequencing platforms have proved to be highly efficient in study of viruses identification and its interactions with host (Mandadi and Scholthof, 2015; Prabha et al., 2013; Rubio et al., 2015; Sharma et al., 2013; Vaucheret, 2006; Wang, 2015). This approach exploits a natural and fundamental antiviral defense mechanism called RNA interference (RNAi). In eukaryotes, upon viral infection RNAi employs Dicer and Dicer-like (DCL) enzymes to cleave viral RNAs into small interfering RNAs (siRNAs) with sizes about 21 nucleotides (nt), which are further amplified by RNAdependent RNA polymerases (RdRPs). These siRNAs are loaded into argonaute to form the RNA induced silencing complex to specifically silence target genes (Baulcombe, 2006; Ding, 2010). Based on the mechanism, Kreuz et al., first proposed that high-throughput small RNA sequencing (sRNA-seq) can be used as a routine method to detect various DNA or RNA viruses (Kreuze et al., 2009). Subsequently, to streamline this technology, Li et al. (2012, 2013, 2015) explored the VirusDetect, a bioinformatics pipeline that can efficiently analyze large scale small RNA (sRNA) datasets for both known and novel virus identification in plants and animals (Li et al., 2012, 2013, 2015; Zheng et al., 2017). Furthermore, Wu et al. (2010) also suggested a possibility of using sRNAs for viral genome assembly and virus discovery in invertebrate, such as fruit fly, mosquito and nematode. Recently, the method was used to detect the human viruses (Wang et al., 2016). In this study, we also identified and characterized the CymMV and ORSV-derived siRNAs and predicted its host target genes from disease P. equestris plants through sRNA-seq to investigate the interaction between these two viruses and its host plant.
Materials and Methods
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted from 0.1 g symptomatic leaves of P. equestris with Trizol reagent (Invitrogen, Carlsbad, CA, USA) as instruction. RT-PCR was performed with the primers to amplify the coat protein (CP) gene of CymMV and ORSV with Invitrogen SuperScript IV VILOTM kit (ThermoFisher, Waltham, MA, USA) as instruction.
Electron microscopy
Infected and healthy P. equestris leaves were investigated for virus particles by both negative staining and thin sections under H-7650 Hitachi transmission electron microscope (Tokyo, Japan) at 80 kV as described (Lan et al., 2016).
sRNA-seq and analyzing
The RNA extracted from infected and healthy P. equestris (n = 3) was used for library construction and sequencing for three times by Novogene Technology Co. Ltd. (Beijing, China) as described previously (Lan et al., 2015). To identify siRNAs, using the software Bowtie v.0.12.7 with 0 mismatch, we aligned all the cleaned reads to the viral genome sequences and calculated the average depths and the genome coverages. The average depth is calculated as the total number of nucleotides of the aligned reads divided by the read-covered positions on the reference genome. The genome coverage represents the proportion of read-covered positions against the genome length. Reads obtained above were analyzed using Excel. Single-base resolution maps along with virus genomes were created using Bowtie tools and in-house Perl scripts. sRNAs from each sample were assembled de novo using Velvet. Assembled contigs were used to search the Gen-Bank/EMBL/DDBJ database using BLASTn or BLASTx. Coverage and distribution of virus-specific contigs by siRNAs were determined using the program MAQ under default parameters, and results were exported to Microsoft Excel for further analysis. Primers were designed for amplification and Sanger sequencing based on the identified viral contigs using Vector NTI (Invitrogen). Phylogenetic tree analysis was performed using MEGA 5.0.
Real-time RT-PCR (RT-qPCR)
We obtained the sequences of the RNAi pathway core components in the genome of P. equestris through a local BLASTx search with corresponding sequences of Oryza sativa as queries (Cai et al., 2015; Kapoor et al., 2008; Niu et al., 2016). Furthermore, we used the sRNA target analysis server psRNATarget (Dai and Zhao, 2011) to predict putative target genes of P. equestris by CymMV- and ORSV-derived siRNAs. We then investigated the transcript levels of these core components and target genes by RT-qPCR assay on a Quant-Studio 6 Flex Real-Time PCR System (Life Technologies, Carlsbad, CA, USA) using the SYBR Green PCR Master-Mix kit (Promega, Madison, WI, USA) with primers (Table 1) as described previously (Lan et al., 2015, 2018a).
Results and Discussion
Identification of CymMV and ORSV infection in P. equestris
P. equestris samples showing severe chlorotic, ringspots even necrotic symptoms (Fig. 1A) caused by CymMV and ORSV infection about 15 days were collected from Orchid Cultivation Distribution Center in Zhangzhou, Fujian. The co-infection of CymMV and ORSV was confirmed by RT-PCR and by H-7650 Hitachi transmission electron microscopy. Our results showed that coat protein genes of CymMV and ORSV were successfully amplified by RT-PCR (Fig. 1B). Furthermore, both negative staining and thin sections suggested that abundant of approximately 300-nm rod-shaped and 500-nm linear virus particles, representing for CymMV and ORSV, respectively, existed in viruses-infected P. equestris plants (Fig. 1C and D).
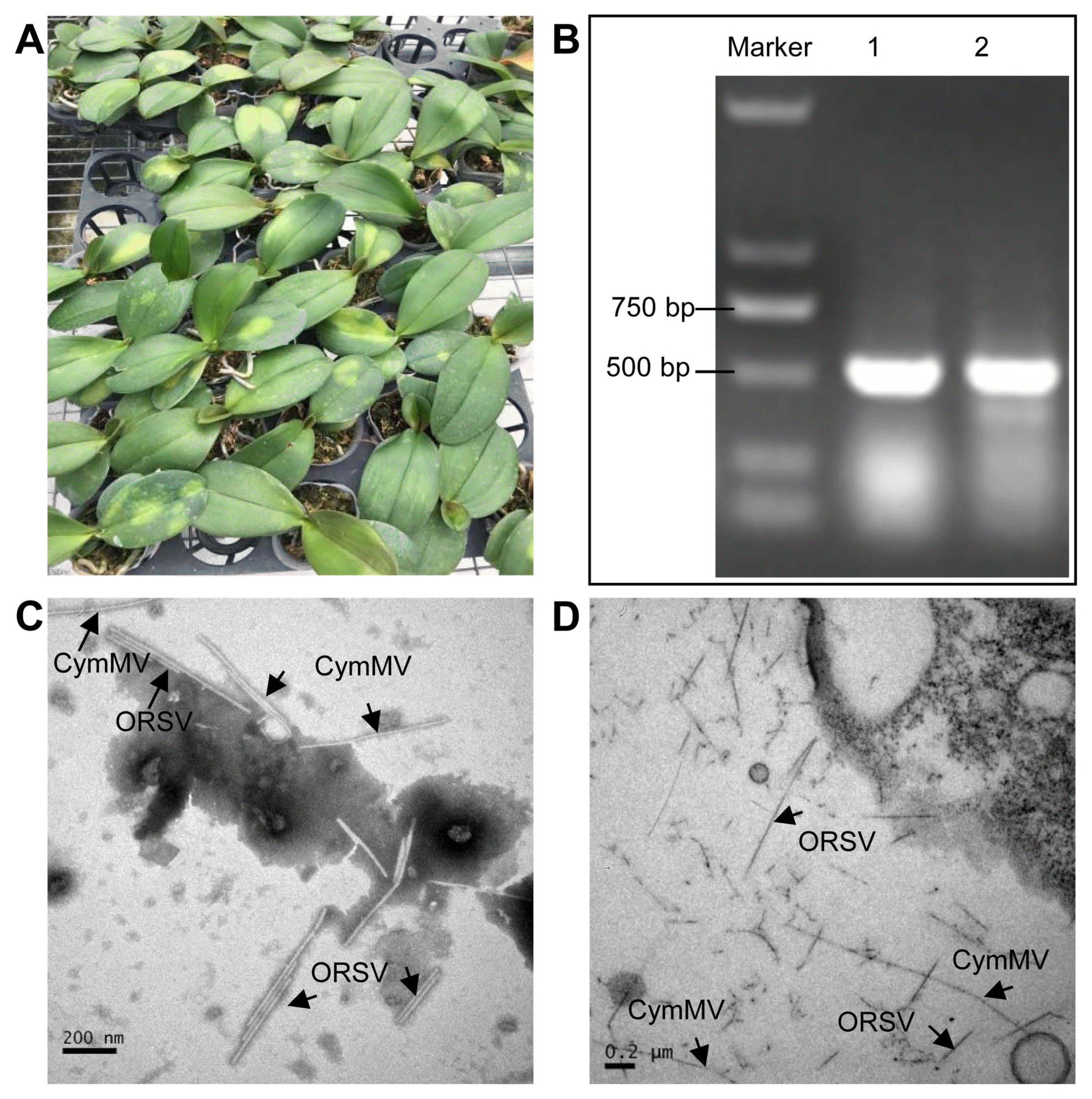
Confirmation of Phalaenopsis equestris co-infected with Cymbidium mosaic virus (CymMV) and Odontoglossum ringspot virus (ORSV) by reverse transcription polymerase chain reaction (RT-PCR) and electron microscopy. (A) Symptoms of Phalaenopsis equestris co-infected with CymMV and ORSV. The infected plant showed severe symptoms on leaves, including chlorotic and necrotic sunken patches. (B) Agarose gel electrophoresis of RT-PCR products of CymMV and ORSV coat protein (CP) gene. M, DNA marker D2000; 1 and 2, reverse transcription polymerase chain reaction products of CymMV and ORSV CP gene, respectively. (C, D) Electron microscopy of Cym-MV and ORSV co-infected P. equestris leaves showing virus particle morphology through the negative staining (C) and thin sections (D). Scale bars = 200 nm.
Characterizations of siRNAs derived from CymMV and ORSV in P. equestris
Following confirmation of CymMV and ORSV co-infection, sRNA from virus-infected and virus-free plant samples was sequenced with the Solexa protocol. Totally, 6,133,689 and 7,563,892 clean reads were obtained from the sRNA libraries of virus-infected and virus-free sample, respectively. Among these reads, a total of 369,524 (6.02%) and 457,104 (7.45%) reads from virus-infected sample were mapped to the CymMV and ORSV genomes, respectively. Conversely, only 1,007 (0.01%) reads matched the two virus genomes in virus-free sample. 21-nt reads increased, while 24-nt reads decreased significantly in the virus-infected sample compared with virus-free sample (Fig. 2A), suggesting that virus infection changed the distribution pattern of sRNA. In virus-infected sample, the majority (72% and 66%) of siRNAs derived from CymMV and ORSV were 18–22 nt in length, with 21 and 22-nt being most abundant (Fig. 2B and C); however, the majority of siRNAs in many virus-plant systems were 21–24 nt in length, with 21- and 22-nt being also most abundant (Kreuze et al., 2009; Lan et al., 2018b; Li et al., 2016; Mitter et al., 2013; Xu and Zhou, 2017; Yan et al., 2010; Yang et al., 2014). Thus, these results implicated that the homologues of DCL4 and DCL2 in P. equestris may be the predominant DCLs involved in the biogenesis of viral small interfering RNAs (vsiRNAs) which functions as the predominant antiviral silencing mediators (Blevins et al., 2006; Deleris et al., 2006; Ding, 2010; Donaire et al., 2008; Liu et al., 2018; Niu et al., 2017); as for other DCLs of P. equestris participating in vsiRNAs generation will be the future discussed topic. Then, we analyzed the features of 21-nt siRNA duplexes induced by CymMV and ORSV. The results showed that 21-nt siRNA duplexes derived from CymMV and ORSV with 0-nt overhangs were the most abundant 21-nt duplexes, followed by 2-nt overhangs and then 1-nt overhangs 21-nt duplexes in P. equestris (Fig. 2D), which was different from that of other viruses-plant and viruses-invertebrates systems, in which the 21-nt siRNAs duplexes with 2-nt overhands were the most abundance duplexes (Liu et al., 2018; Niu et al., 2017). Thus, the results suggested that 21-nt vsiRNAs duplexes with 0-nt overhangs were the most efficient triggers of RNAi in P. equestris. It has been reported that sRNA sequences could be overlapped and further assembled into longer transcript contigs (Kreuze et al., 2009). Our results revealed that 4 and 7 transcript contigs, with a longest contigs of 3,167 and 2,853 nt, were obtained respectively for CymMV and ORSV with the short sequence assembly program Velvet (Fig. 3A and B). The assembled contigs from sRNAs covered about 99% of the CymMV and ORSV genomes (Fig. 3A and B), with an average sequencing depth of 350× and 400×, respectively. RT-PCR and rapid amplification of cDNA ends were respectively used to amplify the gaps between siRNA contigs and terminal sequences using primers designed according to the sequences of Cym-MV and ORSV genomes (data not shown). The obtained PCR fragments were sequenced using the Sanger method and assembled to produce the near complete genomes of both viruses. Phylogenetic analysis using deduced RdRP sequences of CymMV and ORSV confirmed their relatedness to the genus Potexvirus and the genus Tobamovirus, respectively (Supplementary Fig. 1).

Characterizations of siRNAs derived from Cymbidium mosaic virus (CymMV) and Odontoglossum ringspot virus (ORSV) in Phalaenopsis equestris. (A) Size distribution of total small RNAs (18–32 nt) derived from CymMV and ORSV co-infected (black line) and viruses-free (pink line) P. equestris plants. (B, C) Size distribution of total and unique small interfering RNAs (siRNAs; 18 to 32 nt) matching CymMV (B) and ORSV (C) genomes from co-infected P. equestris plants. (D) Reads of 21-nt siRNAs with 1–21 nt distance between 5′ ends in CymMV (red line) and ORSV (black line)-infected P. equestris plants. (E, F) Relative frequency of 5′-terminal nucleotide of total and unique viral small interfering RNAs (vsiRNAs) (18 to 32 nt) matching CymMV (D) and ORSV (E) genomes from co-infected P. equestris plants. (G, H) Polarity distribution of total and unique vsiRNAs (18 to 32 nt) matching CymMV (F) and ORSV (G) genomes from co-infected P. equestris plants. “+” and “−” indicate vsiRNAs derived from positive and negative genomic strands, respectively. The data shown was the representative results of three replicates of small RNA sequencing.
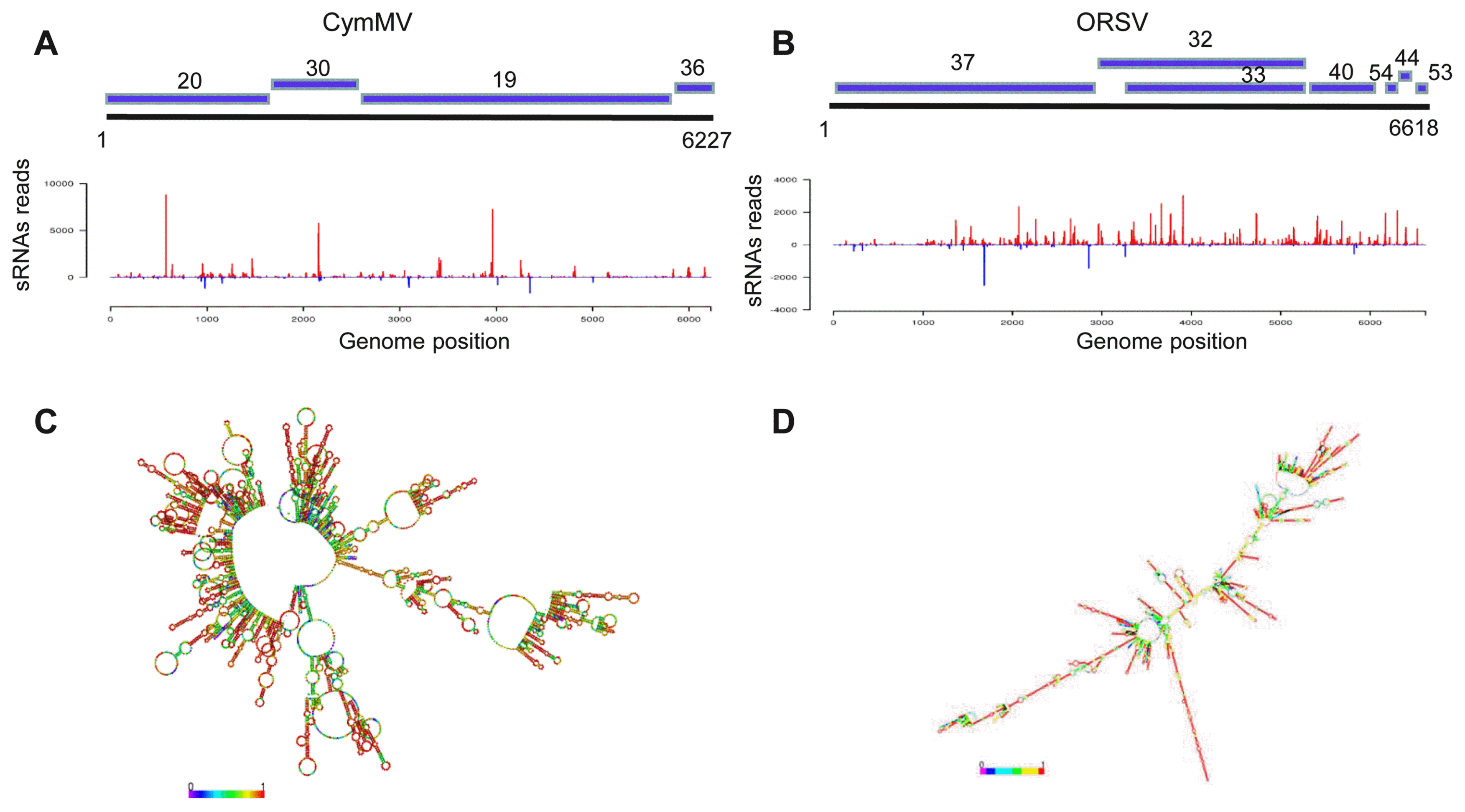
Generation sites of viral small interfering RNAs are associated with the putative secondary structures in the virus genomes RNA of Cymbidium mosaic virus (CymMV) and Odontoglossum ringspot virus (ORSV). (A, B) Coverage of viral genomes by long contigs assembled from small interfering RNAs (siRNAs). Up panel, long contigs assembled from siRNAs. Down panel, distribution and hotspots for siRNAs along respective genomes. sRNA, small RNA. (C, D) Secondary structures of CymMV and ORSV RNAs predicted with RNAfold server (http://rna.tbi.univie.ac.at/). Color bar with No. 0–1 denotes the possibilities of bases paring. The data shown was the representative results of three replicates of small RNA sequencing.
Previous studies have reported that the 5′-terminal nucleotide of sRNAs play irreplaceable roles in mediating the sorting of sRNAs into AGO complexes in plants (Mi et al., 2008). Consistent with previous report for diverse plant-virus-specific sRNAs (Donaire et al., 2008; Lan et al., 2018b; Xu and Zhou, 2017), CymMV and ORSV siRNAs also demonstrated a clear tendency to begin with uracil (U), adenine (A), as compared with cytosine (C) and guanidine (G) (Fig. 2E and F). To explore the origin of vsiRNAs, their polarity distribution was further characterized. Our results showed that CymMV and ORSV siRNAs were derived predominantly from the viral positive-strand RNA, accounting for about 90% (Fig. 2G and H). This is broadly contrasted to many single-stranded RNA (ssRNA) positive-strand viruses, siRNAs of which were produced equally from the positive and the negative strands or predominantly from negative strands of viral genomes (Ho et al., 2007; Lan et al., 2018b; Li et al., 2016; Xia et al., 2014; Yang et al., 2014). Furthermore, single-base resolution maps of total CymMV and ORSV siRNAs along with genomes were created using Bowtie tools and in-house Perl scripts. Our results revealed that majority siRNAs were from the intergenic regions, not the 5′- or 3′-terminal regions of the two viruses genomes and siRNAs had a continuous but heterogeneous (Hot spot and Cold spot) distribution along the genomes of the two viruses (Fig. 3A and B). Taken together, our results implied that highly structured regions present in CymMV and ORSV genomes may be the substrates for DCL enzymes. To explore the possibility, we evaluated the secondary structures of CymMV and ORSV with RNAfold server. Our results showed that lots of stem-loop secondary structures were formed in intergenic regions and corresponded to the hotspots (Fig. 3C and D). Taken together, these results supports that siRNAs should originate predominantly by direct DCL cleavage of imperfect duplexes in the most folded regions of the positive strand of both viruses RNA molecular in P. equestris, similar to the speculation for siRNAs origin from plant infected with Cymbidium ringspot tombusvirus (CymRSV), a ssRNA positive-strand virus, implying siRNAs in the CymRSV system are also predominantly processed by DCL cleavage of imperfect duplexes, which can be formed from the positive-strand viral RNA (Molnár et al., 2005).
siRNAs generated by RNAi response down-modulated the expression levels of endogenous genes in P. equestris
To further confirm the response of RNAi pathway to CymMV and ORSV infection, we obtained the sequences of the RNAi pathway core components in the genome of P. equestris through a local BLASTz search with corresponding sequences of O. sativa as queries (Cai et al., 2015; Kapoor et al., 2008; Niu et al., 2016). We then investigated the transcript levels of these core components by RT-qPCR assay on a QuantStudio 6 Flex Real-Time PCR System (Life Technologies) using the SYBR Green PCR MasterMix kit (Promega) as described previously (Lan et al., 2015, 2018a). In total, we identified 14 core components of RNAi pathway in P. equestris genome, including DCL1, DCL2a, DCL3a, DCL4, AGO4B, AGO7, AGO10, AGO16, AGO-PNH1, AGO-MEL1, RdRP1, RdRP2, RdRP5, and RdRPSHL2 (Supplementary Fig. 2). As indicated in Fig. 4, RT-qPCR assays demonstrated that DCL2a, DCL4, AGO4B, AGO7, AGO10, RdRP1, and RdRP5 transcript levels increased significantly by 2–6 folds (Fig. 4); in contrast, transcript levels of DCL1, AGO16, and RdRP-SHL2 decreased obviously by 50–70% (Fig. 4) in virus-infected P. equestris compared with virus-free P. equestris. Transcript levels of other genes of RNAi pathway change slightly (Fig. 4). Researches have shown that down-regulation of DCL2 and DCL4 in virus-infected rice may influence the production of 21- and 22-nt vsiRNAs and thus weaken host defense, allowing viruses to establish successful infection. For example, mutation of DCL4 from O. sativa can induce abnormal expression of auxin response factors and lead to growth abnormities (Liu et al., 2007). In our study, however, DCL2a and DCL4 were up-regulated but DCL1 was down-regulated (Fig. 4). Thus, the down-regulation of DCL1 in virus-infected P. equestris might partially account for P. equestris symptoms showing severe chlorotic, ringspots even necrotic. DCL2a and DCL4 may have additional biological pathways or functions. Many viral suppressors of RNA silencing (VSRs) can block biogenesis of vsiRNAs by interacting with plant DCLs. For example, DCL4 from O. sativa is suppressed by RYMV P1 (Lacombe et al., 2010). It is yet to confirm whether VSR proteins encoded by CymMV and ORSV interacts with the DCL1 of P. equestris. Plant AGOs participate in host defense against many viruses species. In our study, CymMV and ORSV infection-induced up-regulation of AGO4B, AGO7, and AGO10 but down-regulation of AGO16 (which was predicted as a target of a certain vsiRNA, as after-mentioned) (Fig. 4), and in particular, AGO4B and AGO7 was drastically up-regulated (Fig. 4), suggesting facilitated host antiviral defense. The dramatically activated AGO4B and AGO7 might not only act in vsiRNAs binding, but also have additional biological functions. Whether the down-regulated AGO16 in virus-infected plant affected RNA silencing–directed antiviral defense is unclear and would be the topic in future. RdRP6 from O. sativa plays role in defense against rice viruses (Hong et al., 2015; Jiang et al., 2012). In our study, RdRP5 and RdRP1 were up-regulated in infected P. equestris, implying their counterpart roles in host response to CymMV and ORSV infection. In general, these results suggested the complicated interactions between CymMV/ORSV and P. equestris RNA silencing pathway, which likely are modulated by the intertwined effects of multiple RNAi components.
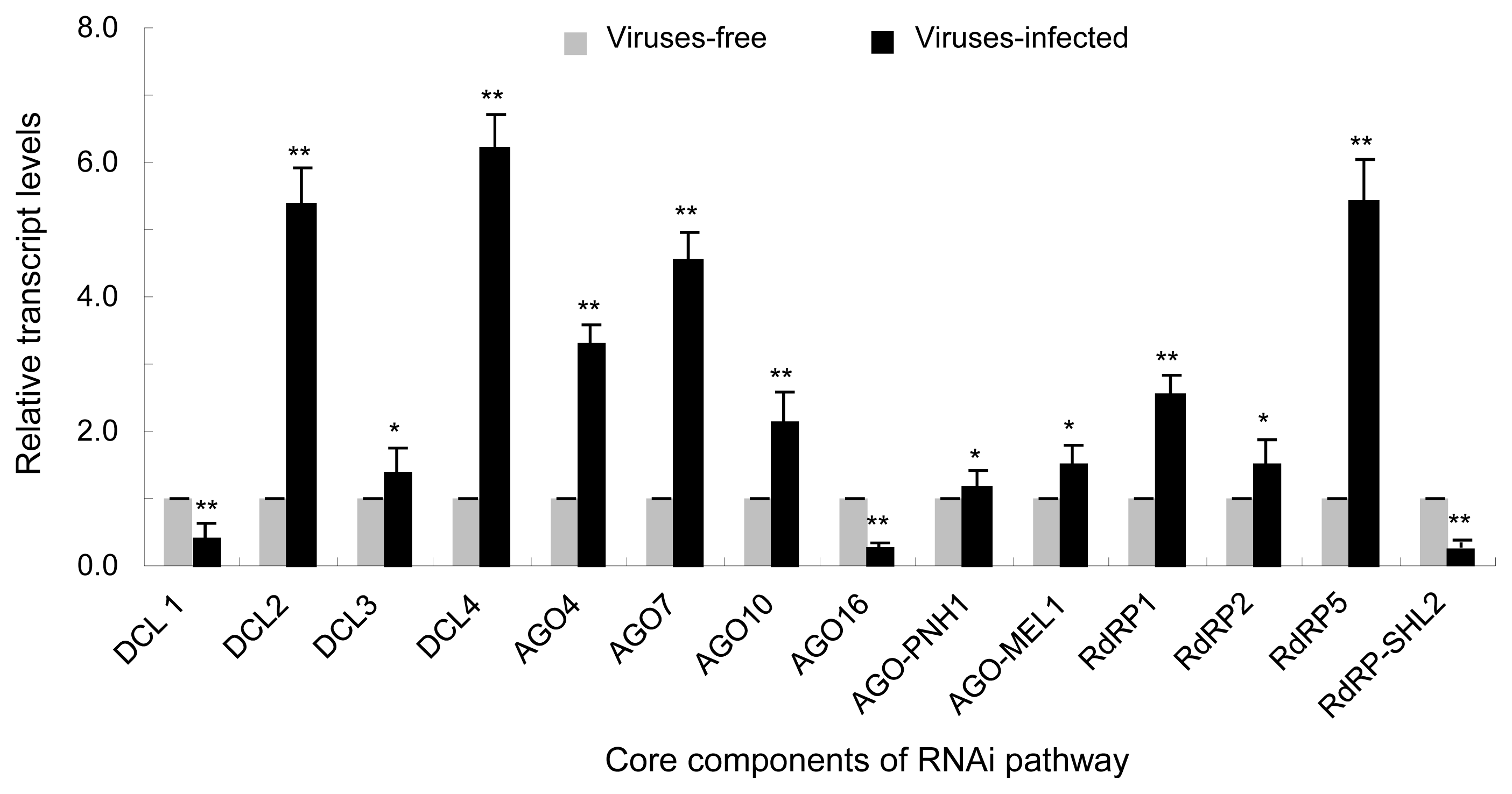
Core components of RNA interference (RNAi) were selectively and differently modulated by infection of Cymbidium mosaic virus (CymMV) and Odontoglossum ringspot virus (ORSV) in Phalaenopsis equestris. Relative transcript levels of core components of RNAi in CymMV and ORSV co-infected and viruses-free P. equestris as detected by real-time reverse transcription polymerase chain reaction assay with mean ± standard deviation (SD) of three independent experiments. Statistical analysis was conducted based on Tukey’s honest significant difference test using SAS version 4 (SAS Institute, Cary, NC, USA). **P < 0.01, *P < 0.05. DCL, Dicer-like; RdRP, RNA-dependent RNA polymerase.
Target identification is very important to understand deeply the functions and implications of viral siRNAs in its interaction with host. In this study, we used the sRNA target analysis server psRNATarget (Dai and Zhao, 2011) to predict putative target genes of P. equestris by CymMV- and ORSV-derived siRNAs. Because of the large number of putative genes targeted by siRNAs, only those gene sequences with more than 12 continuous base-pairing with vsiRNAs were selected for subsequent analysis. In total, 21 and 33 putative target genes were predicted by CymMV- and ORSV-derived siRNAs, respectively (Table 2). Annotation of the 54 genes predicted was done referring to the De novo transcriptome assembly databases in the P. equestris (Niu et al., 2016). Among of those annotated target genes, many were related to disease/stress response, including nine R genes which were annotated as NBS-LRR or NBS protein genes, one F-box/LRR or F-box domain-containing protein gene, three ubiquitination-related protein genes, four receptor-like protein kinase (RLK) genes, and one universal stress protein (USP) gene (Table 2). A majority of NBS-LRR protein genes play important roles in pathogen recognition and host defense against a variety of pathogens including viruses, bacteria and fungi (DeYoung and Innes, 2006; McHale et al., 2006). Plant F-box proteins are involved in stress responses. It also has been shown that F-box proteins and RNAi pathway could modulate each other in plants; even more, some plant viruses encode an F-box-containing viral silencing suppressors (VSRs) to down-regulated RNAi pathway through the autophagy (Derrien et al., 2012; Earley et al., 2010; Jones-Rhoades et al., 2006; Kim and Delaney, 2002). RLKs also participate in pathogen recognition and resistance (Morris and Walker, 2003). USPs can increase adversity tolerance of plants (Sinha et al., 2016). Besides, our results revealed that 4 target genes (DCL1, argonaute16, RdRP-SHL2, and piRNA biogenesis protein EXD1) were annotated as core components of RNAi response (Table 2), suggesting RNAi response may be targeted and its expression levels be further suppressed by vsiRNAs generated in P. equestris infected with CymMV and ORSV, which is similar to other host-pathogen systems (Xu and Zhou, 2017). Many intracellular pathogens, including viruses, parasites, and bacteria, invade, replicate, spread within and exit their host cells in a cytoskeleton-dependent manner (Gouin et al., 2005; Greber and Way, 2006; Gruenheid and Finlay, 2003). In this study, 9 target genes (mainly including myosin, networked 1A, pentatricopeptide repeat-containing protein, and vimentin) were annotated as cytoskeleton-related proteins (Table 2), implying they play important roles in CymMV and ORSV infections in P. equestris. Moreover, several chloroplastor mitochondria-related genes were annotated (Table 2), implying that photosynthesis and energy supply pathways might be influenced by CymMV and ORSV infections P. equestris through vsiRNAs targeting.
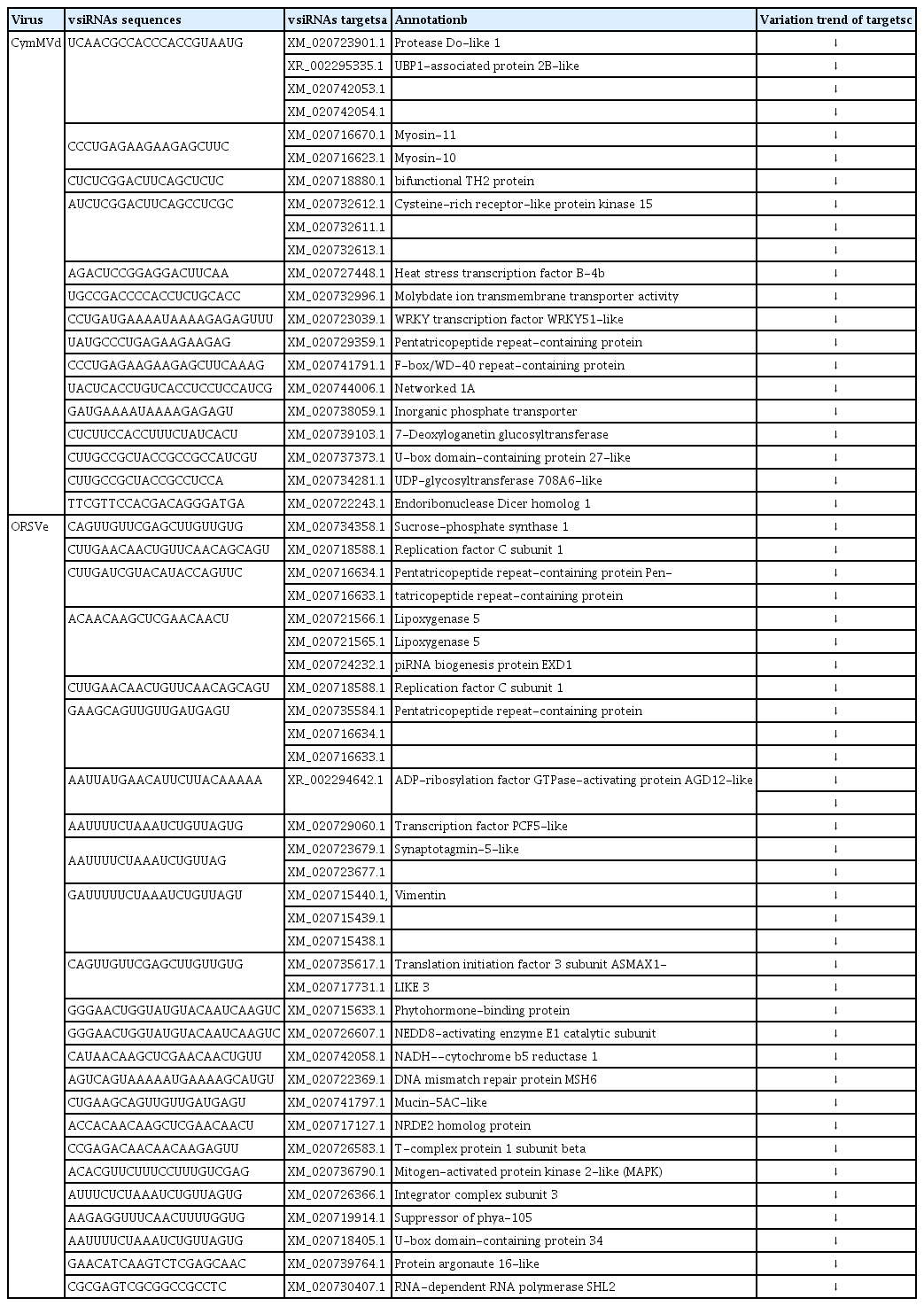
Functional annotations and variation trends of the predicted target genes of siRNAs derived from CymMV and ORSV in Phalaenopsis equestris
In BLAST2GO analysis, a total of 54 predicted target genes of CymMV and ORSV siRNAs were annotated and classified into three Gene Ontology (GO) categories (biological process, cellular component, and molecular function) (Fig. 5). For target genes of CymMV siRNAs, five terms under the biological process section, “stress response,” “defense response or innate immune responses,” “protein phosphorylation,” “metabolic process,” and “cytoskeleton,” represented 85% of the classified targets (Fig. 5A); the four most highly represented GO terms were “integral component of membrane,” “nucleus,” “myosin complex,” and “proteasome” under the cellular component category, comprising 75% of the annotated genes (Fig. 5A); and the five most abundant GO terms under the molecular function category, “protein kinase activity,” “nucleic acid binding,” “ubiquitin-protein transferase activity,” “DNA binding transcription factor activity,” and “microtubule motor activity,” represented 65% of the annotated genes (Fig. 5A). For target genes of ORSV siRNAs, four terms under the biological process section, “fatty acid biosynthetic process,” “RNAi,” “cytoskeleton,” and “DNA replication,” represented 86% of the classified targets (Fig. 5B); the four most highly represented GO terms were “RNAi components,” “intermediate filament,” “integral component of membrane,” and “DNA replication factor C complex” under the cellular component category, comprising 60% of the annotated genes (Fig. 5B); and the seven most abundant GO terms under the molecular function category, “binding”, “oxidoreductase activity,” “siRNA metabolic process,” “structural molecule activity,” “hydrolase activity,” “regulation of transcription,” and “protein kinase activity”, represented 83% of the annotated genes (Fig. 5B). Thus, there are some similar and different GO categories when compared with target genes of CymMV siRNAs (Fig. 5B).

Gene Ontology categories of predicted target genes of small interfering RNAs (siRNAs) derived from Cymbidium mosaic virus (CymMV) and Odontoglossum ringspot virus (ORSV) in Phalaenopsis equestris. The predicted target genes were subject to BLAST alignment and Gene Ontology annotation using BLAST2GO software. A and B denoted respectively the predicted genes targeted by siRNAs derived from CymMV and ORSV in P. equestris. RNAi, RNA interference.
To confirm whether the predicted target genes were modulated by siRNAs, we conducted RT-qPCR to detect all target genes expression levels of P. equestris. The result showed that all the target genes were down-regulated with different degree and 15 of them had significantly decreased expression levels in the CymMV and ORSV infected plants compared to the uninfected control (Table 2, Fig. 6). In other pathogen host systems, viral siRNAs also were shown to target and down modulate the expression levels of its corresponding target genes in host (Lan et al., 2018b; Xu et al., 2017). Thus, these results suggested the potential roles of viral siRNAs in modulating growth and development of host.
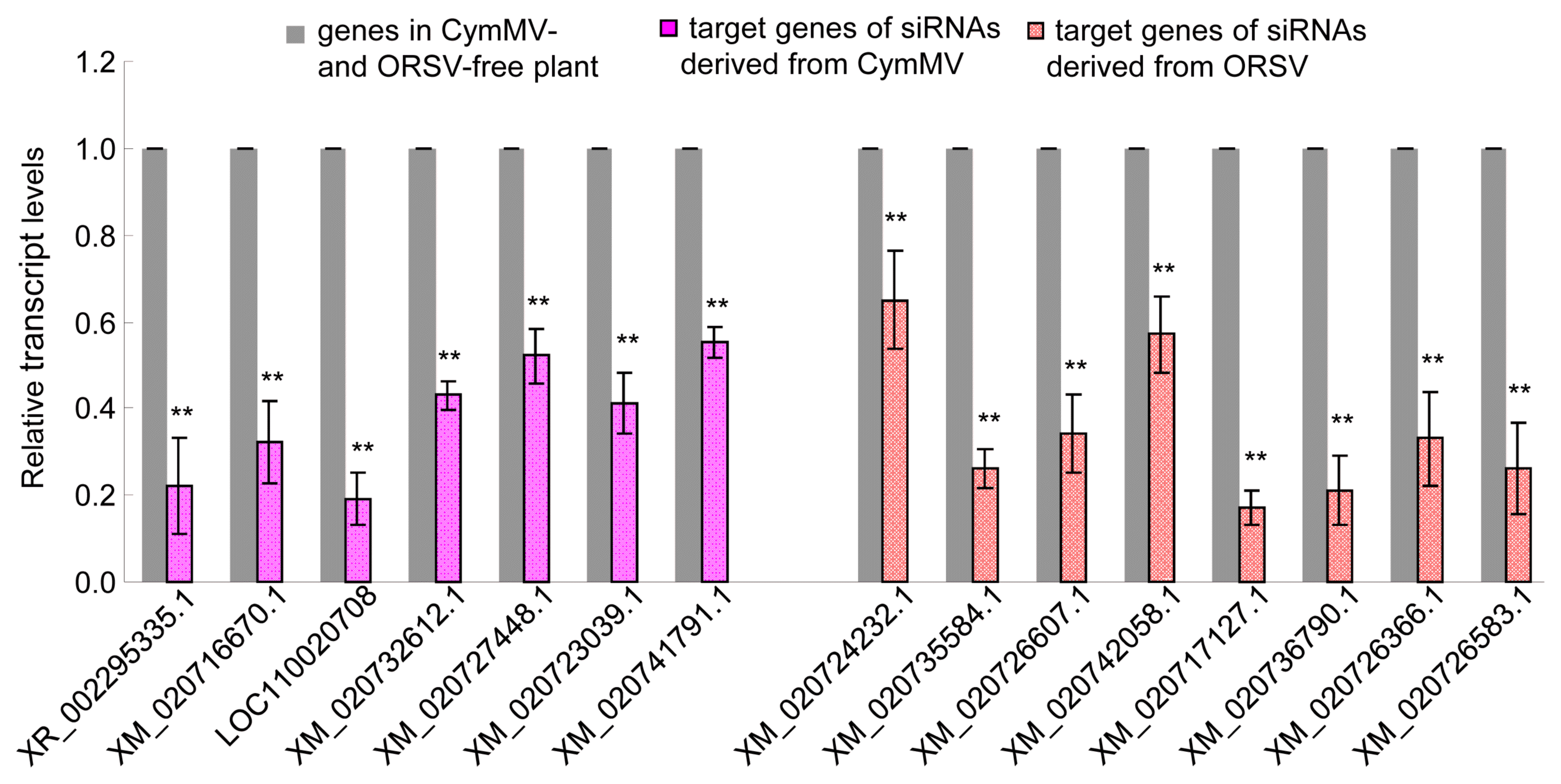
The expression levels of partial predicted target genes were down-regulated by small interfering RNAs (siRNAs) derived from Cymbidium mosaic virus (CymMV) and Odontoglossum ringspot virus (ORSV) in Phalaenopsis equestris. The expression levels were detected by real-time reverse transcription polymerase chain reaction assay with mean ± standard deviation (SD) of three independent experiments. Statistical analysis was conducted based on Tukey’s honest significant difference test using SAS version 4 (SAS Institute, Cary, NC, USA). **P < 0.01.
In this study, we analyzed the characterizations of the siRNAs in CymMV- and ORSV-coinfected P. equestris plants, and explored their potential roles in targeting a series of P. equestris genes which are related to host stress response, RNAi antiviral immunity defense, cytoskeleton, metabolic processes (chloroplast photosynthesis and mitochondria energy supply pathways, fatty acid biosynthetic process and protein phosphorylation), pathogenesis and symptomology. We also found that siRNAs derived from CymMV and ORSV could decrease the expression levels of all target genes in P. equestris. All results in this study provide an understanding of the RNAi mechanism and an insight into the function and implication of RNAi pathway in viruses-P. equestris interactions.
Supplemental Materials
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no. 31601613), the Nature Science Foundation of Zhangzhou (grant no. ZZ2017J03), the Nature Science Foundation of Fujian (2018J01465) and the Outstanding Youth Research Talent Project for Fujian University (2017).
Notes
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
