Taxonomic Re-evaluation of Colletotrichum gloeosporioides Isolated from Strawberry in Korea
Article information
Abstract
For the past two decades, the causal agent of anthracnose occurring on strawberry in Korea was considered Colletotrichum gloeosporioides. However, the recent molecular analysis has shown that the genus Colletotrichum has undergone many taxonomic changes with introduction of several new species. As a result, it revealed that C. gloeosporioides indeed consisted of more than 20 distinct species. Therefore, the Korean pathogen isolated from strawberry should be reclassified. The shape and size of the conidia of the pathogen were not distinctly different from those of C. gloeosporioides and C. fructicola, but it differed in shape of the appressoria. A combined sequence analysis of partial actin, glycer-aldehydes-3-phosphate dehydrogenase genes, and the internal transcribed spacer regions showed that the strawberry isolates formed a monophyletic group with authentic strains of C. fructicola. On the basis of these results, the anthracnose fungi of the domestic strawberry in Korea were identified as C. fructicola and distinguished from C. gloeosporioides.
Anthracnose crown rot, to which all currently grown strawberry (Fragaria × ananassa Duch.) cultivars are susceptible, is a major disease in over 30% of nurseries and after transplanting stage in Korea (Nam et al., 2009). The causal pathogen Colletotrichum gloeosporioides was first reported by Kim et al. (1992).
C. gloeosporioides that occurs in many crops is a species aggregate containing a number of polymorphic subgroups which show varying degrees of pathogenicity, host-specificity and genetic homogeneity (Hyde et al., 2009a). However, the recent epitypification of C. gloeosporioides has enabled accurate classification by comparative analysis with pathogens isolated from the various plants (Cannon et al., 2008). The sequence analysis of the type strains was an important element in the research of identification and phylogenetic relationship of Colletotrichum (Cai et al., 2009; Phoulivong et al., 2010). Anthracnose pathogens, previously reported to be Colletotrichum sp., that occur on coffee in Thailand were shown as three new species of C. asianum, C. fructicola, and C. siamense based on the multigene sequence analysis and morphological characteristics (Prihastuti et al., 2009). In the past few years, anthracnose pathogens from tropical fruits, which were identified as C. gloeosporioides based on their morphological characteristics, are subdivided into C. asianum, C. fructicola, C. horii, C. kahawae, and C. gloeosporioides by comparing of the nucleotide sequences with that of the C. gloeosporioides epitype (Phoulivong et al., 2010). As a result, many species of Colletotrichum, including C. gloeosporioides, have been defined, mainly based on the results of molecular phylogenetic analysis (Damm et al., 2010). Although the internal transcribed spacer (ITS) sequence do not separate C. gloeosporioides complex, some single genes or combinations of genes, such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and glutamine synthetase, can be used to reliably distinguish most taxa (Weir et al., 2012).
Therefore, C. gloeosporioides, previously reported pathogen on strawberries in Korea, is necessary to re-evaluate, taking into account the changed species boundary and the recent phylogenetic results of the genus Colletotrichum.
Isolation and culture of strains
Eleven strains isolated from strawberry plants from 2005 to 2011 (Table 1). Diseased petiole, roots, and crown tissues were surface sterilized by dipping in 2% NaOCl for 2 min, rinsed thrice with sterile water, and dried on sterile tissue paper. Samples were then placed on water agar and incubated at 27°C. The growing edges of any fungal hyphae developing from the tissue were then transferred aseptically to potato dextrose agar (PDA; Difco, Detroit, MI, USA). Single spore isolations were also carried out using the procedure described by Prihastuti et al. (2009). Pure cultures were stored at 4 °C on PDA slants.
Pathogenicity test
Each isolated pathogen was prepared at 1 × 105 conidia/mL and spayed 1 mL per plant on the Seolhyang cultivar of strawberry plants. Six plants per an isolate were used for test. The inoculated plants were incubated in a dew plastic house at 27 °C and 100% relative humidity (RH) for 2 days and then moved to a plastic house held between 24 °C and 30 °C. After 60 days, disease index on each plant was rated on a scale of 0–4: 0, healthy; 1, < 50% of petioles affected; 2, > 50% of petioles affected; 3, wilted; 4, necrosis formed on the entire plant (Nam et al., 2006). Disease index data from isolates were subjected to analysis of means and standard error. All data analysis was performed using the Costat program (CoHort software, Berkeley, CA). All isolates caused anthracnose symptoms on the Seolhyang strawberry cultivar (Table 1). Disease index of the isolates was the highest (DI > 3) for CGF071204, CGF100713, and CGF110101. Smith and Black (1990) showed that pathogenicity of C. gloeosporioides, C. acutatum, and C. fragariae can be differentiated by factors such as strawberry tissue and cultivars. C. gloeosporioides and C. acutatum were reported to have a wide host range, while C. fragariae were restricted to strawberry plants (Freeman et al., 1998; Hyde et al., 2009b). Colletotrichum isolated from strawberry in Korea and Japan was pathogenic to strawberry and apple fruit (Nam et al., 1998), and avocado, broad bean, common sowthistle, cyclamen, pea, and strawberry under non-wound inoculation condition (Okayama and Tsujimoto, 1994), respectively.
Morphological and cultural characteristics
Isolates were measured for colony characteristics, and shape and size of the conidia after 7 days of incubation on PDA media. The PDA first appeared as white, then becoming grey to dark colony diameters were measured daily for 7 days, and the grey at the center over time, and growth rate was shown growth rate for these 7 days was calculated as the average 9.98 ± 1.8 mm per day (Table 2). The shape and size of the of mean daily growth (mm per day). The colors of the conidia were cylindrical and 14.6 ± 0.47 ×3.99 ± 0.11 μm, conidial masses and zonation were recorded at 7 days. respectively. The shape and size of the appressoria were Appressoria were produced using a slide culture technique, ovoid, sometimes clavate and 9.93 ± 0.32 ×4.90 ± 0.10 μm, in which a conidial suspension was placed in a hole on the respectively (Table 2). Prihastuti et al. (2009) reported that glass slide and covered with a cover slip. The slide was the colony characteristics and conidial shape and size of C. placed in an empty Petri dish with wet filter paper and fructicola were similar to those of C. gloeosporioides, but incubated at 28 °C for 2–4 days. Colonies of the isolates on that ovoid, sometimes clavate appressoria in C. fructicola were different from circular to slightly irregular one in the latter species. Size of conidia and appressoria of the strawberry isolate differed from C. fragariae by Gunnell and Gubler (1992). Therefore, Colletotrichum isolates from strawberries in Korea were similar to C. gloeosporioides and C. fructicola in shape and size of conidia, but the shape of the appressoria was close to C. fructicola rather than C. gloeosporioides.
Phylogenetic analysis
The sequences of eleven strains from strawberry and the ex-type or authentic strains of anthracnose available from GenBank (Table 3) were used in this study. Genomic DNA was extracted by using the method of Park et al. (2005). For the amplification of the ITS, partial actin (ACT), and GAPDH genes, 3 different primer sets were used: ITS5 and ITS4 (White et al., 1990); ACT512F and ACT783R (Prihastuti et al., 2009); and GDF1 and GDR1 (Peres et al., 2008); respectively. The amplification was performed with Maxime polymerase chain reaction (PCR) PreMix (I-Taq, iNtRoN BioTechnology, Korea) in a final volume of 20 μl containing 10 pmol of each primer set and PCR conditions were performed as described by Prihastuti et al. (2009). PCR products were electrophoresed through a 1% agarose gel stained with ethidium bromide and purified using a PCR quick-spin PCR product purification kit (iNtRON BioTechnology, Korea) according to the manufacturer’s instructions. The nucleotide sequences were determined by Bioneer Corporation (Chungwon, Korea). The sequences were proofread, edited, and merged onto comparable sequences using the PHYDIT program version 3.2 (Chun, 1995). To determine the phylogenetic positions of the Colletotrichum isolates, the alignments of the 3 gene sequence of 11 Korean isolates and 34 reference sequences retrieved from GenBank were performed with Clustal × 1.81 (Thompson et al., 1997). Nucleotide sequences were aligned manually where necessary. A maximum likelihood tree was inferred in MEGA v. 5.05, using TN93+I that was selected as the most suitable nucleotide mode by Modeltest (Tamura et al., 2011). Bootstrap analysis was performed with 1,000 replications for branch stability. Sequences of ITS, ACT, and GAPDH regions of three isolates CGF080901, CGF090501, and CGF100713 were deposited in GenBank (JX125050, JX125051, and JX125052; KC283023, KC283024, and KC283025; KC283017, KC283018, and KC283019, respectively).
PCR amplification of the ITS region generated 466 bp fragments for all isolates from strawberry plants. Sequence analysis of ITS region showed no sequence difference among all isolates and authentic of C. fructicola (ICMP 18581, ICMP 18646, and ICMP 17921) (data not shown).
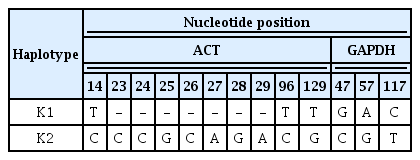
PCR amplification of the ACT gene generated 217–226 bp fragments for all isolates from strawberry plants. Korean isolates present two haplotypes (K1 and K2) (Table 1). The ACT sequence of K1 (CGF050604 and CGF060617) differed from those of other nine Korean isolates (K2) by 3nt polymorphic characters at 14, 96, and 129 position and 7nt deletion at 23–29 position (Table 4). ACT sequence of K1 is identical to that of C. fructicola strain ICMP 17921 (syn. Glomerella cingulate var. minor), whereas K2 showed a sequence similarity of 99.4% with C. fructicola ICMP 18646 (syn. C. ignotum).
PCR amplification of the GAPDH genes generated 232 bp fragments for all isolates. K1 differed from those of other Korean isolates (K2) by 3nt polymorphic characters at 47, 57 and 117 position (Table 4). GAPDH sequence of K1 showed a sequence similarity of 99.5% with that of C. fructicola strain ICMP 17921, whereas K2 showed a sequence similarity of 99.1% with C. fructicola ICMP 18581 and C. fructicola ICMP 18646.
The combined analysis showed that the 11 strains isolated from Korea formed a monophyletic group with authentic of C. fructicola (ICMP 18581, ICMP 18646, and ICMP 17921) and clearly distinct from other C. gloeosporioides complex (Fig. 1). All isolates were divided into two subgroups: one subgroup (K1) contained two Korean isolates and C. fructicola strain ICMP 17921 with a bootstrap value of 77%. The other subgroup (K2) contained 9 Korean isolates and C. fructicola ICMP 18581 and C. fructicola ICMP 18646 with a bootstrap value of 63%.

The maximum-likelihood tree inferred from a combined dataset of ACT, GAPDH and ITS sequences showing phylogenetic relationships among Colletotrichum from strawberry plants in Korea and representative species of C. gloeosporioides complex. Numbers above each branch represent bootstrap values obtained after a bootstrap test with 1,000 replications. Bar indicates the number of nucleotide substitutions.
Since C. fructicola was first isolated from the berries of Coffea arabica in the Chiang Mai region of Thailand (Prihastuti et al., 2009), it has been found in tropical fruits such as chili, papaya, and longan (Phoulivong et al., 2010). Recently, G. cingulata var. minor and C. ignotum placed in synonymy with C. fructicola (Weir et al., 2012). K1 was more closely related to C. fructicola strain ICMP 17921 (syn. G. cingulata var. minor) from Ficus than K2, whereas K2 grouped to C. fructicola ICMP 18581 and C. fructicola ICMP 18646 (syn. C. ignotum).
The Colletotrichum species isolated from strawberries worldwide are as follows: C. gloeosporioides (Gunnell and Gubler, 1992), C. fragariae (Smith et al., 1990), C. acutatum (Smith etal.,1986), C. dematium (Beraha and Wright, 1973), and Gloeosporium spp. (Wright et al., 1960), but as C. gloeosporioides (perfect stage: G. cingulata) in Korea (Kim et al., 1992; Nam et al., 1998). Three anthracnose stains (KACC 40695, KACC 40696, and KACC 40812) from strawberry plants stored in Korean Agricultural Culture Collection (KACC) were identified as C. gloeosporioides based on sequence analysis of ITS and partial β-tubulin gene and cultural characteristics (Kim et al., 2006). In ITS analysis, KACC 40695 and KACC 40812 formed the monophyletic group with K2 and showed 100% sequence similarity among K2, while KACC 40696 was closely related to C. aenigma strain ICMP 18608, C. alienum strain ICMP 12071, and C. siamense ICMP 18578 (syn. C. hymenocallidis) and differed from ex-types of C. fructicola (data not shown). As we performed comparative analysis with three KACC strains using ITS sequence in the present study, phylogenetic analysis with combined genes will be needed to establish for accurate identification of the three stains. The C. fragariae, originally reported as a disease of strawberry in Florida, USA (Brooks, 1931), placed in synonymy with C. theobromicola (Weir et al., 2012). All isolates from Korea were clearly distinguished from C. theobromicola (syn. C. fragariae) based on morphological and molecular phylogenetic analysis.
In the present study, anthracnose fungi that occur in the domestic strawberry were identified as C. fructicola and distinguished from C. gloeosporioides based on their morphological and molecular phylogenetic features.
Acknowledgements
This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ90703904)” Rural Development Administration, Republic of Korea.