 |
 |
| Plant Pathol J > Volume 39(4); 2023 > Article |
|
Abstract
In plant-pathogen interactions, Magnaporthe oryzae causes blast disease on more than 50 species of 14 monocot plants, including important crops such as rice, millet, and most 15 recently wheat. M. oryzae is a model fungus for studying plant-microbe interaction, and the main source for fungal pathogenesis in the field. Here we report that MoJMJD6 is required for conidium germination and appressorium formation in M. oryzae. We obtained MoJMJD6 mutants (ΔMojmjd6) using a target gene replacement strategy. The MoJMD6 deletion mutants were delayed for conidium germination, glycogen, and lipid droplets utilization and consequently had decreased virulence. In the ΔMojmjd6 null mutants, global histone methyltransferase modifications (H3K4me3, H3K9me3, H3K27me3, and H3K36me2/3) of the genome were unaffected. Taken together, our results indicated that MoJMJD6 function as a nuclear protein which plays an important role in conidium germination and appressorium formation in the M. oryzae. Our work provides insights into MoJMJD6-mediated regulation in the early stage of pathogenesis in plant fungi.
Magnaporthe oryzae is the causal agent of rice blast disease and the most common pathogen of cultivated rice. It has been developed as a model system with which to study fungal-plant interactions, and many genes for virulence and specialized infection structures have been identified (Valent, 1990; Xu et al., 2007). The M. oryzae has a complicated life cycle, including conidiogenesis, conidial germination, appressorium formation, and infective hyphae growth, which provides substantial biological information in fungal pathogenesis. During the biotrophic stage of infection, pyriform conidia play a crucial role in the disease cycle of M. oryzae (Xu et al., 2007). When M. oryzae attached to the plant surface, pyriform conidia begin to germinate, a specialized infection structure known as appressorium then differentiates at the tip of the germ tube (Valent et al., 1991). Mature appressorium can accumulate turgor pressure, providing sufficient force to penetrate the strong leaf cuticle and enter into the plant cells (Kankanala et al., 2007). Then the fungus develops invasive hyphae in the rice leaf and typical necrotic lesions develop on the leaf surface (Pinnschmidt et al., 1995). M. oryzae is a devastating pathogen of rice and wheat. It is a hemibiotroph that exhibits symptomless biotrophic growth for the first 4 to 5 days of infection of susceptible cultivars before becoming necrotrophic.
Recently, epigenetics refers to the heritable changes in gene function due to changes in DNA sequence. These epigenetic changes include DNA methylation, histone modifications, and nucleosome remodeling (Huang et al., 2016; Yamane et al., 2006). Eventually, numerous studies had demonstrated the involvement of JmjC domain-containing demethylases in development, diseases, and organogenesis in human, animals, plants, and plant pathogenic fungi (Huang et al., 2016; Klose et al., 2006; Mosammaparast and Shi, 2010; Yamane et al., 2006). In humans, mutations, deletions, or amplifications of other histone demethylase genes have also been associated with cancer development and aggressiveness, and LSD1, FBXL10, JMJD2A, JMJD2B, JMDJ2C, and JARID1B demethylases have been suggested as possible targets for cancer therapy (Kooistra and Helin, 2012). In addition, JMJD3 is also associated with lung and liver cancers and some hematological malignancies (Agger et al., 2009). The expression of the H3K27 demethylase JMJD3 is induced upon activation of the RAS-RAF signaling pathway. JMJD3 is recruited to the INK4A-ARF locus and contributes to the transcriptional activation of p16INK4A in human diploid fibroblasts (Agger et al., 2009). In Drosophila melanogaster, maternal characterize histone demethylase activity of the entire family of JmjC+N protein Lid (little imaginal discs) was identified (Lloret-Llinares et al., 2008). It shows that Lid is structurally homologous to JARID1, a demethylates H3K4me3 (Lloret-Llinares et al., 2008). In plants, the demethylases of the JmjC domain also play an important role in plant growth, development, and flowering. For example, AtJMJ16 and AtJMJ17 are related to leaf senescence and drought stress response (Huang et al., 2019; Liu et al., 2019). JMJ706 affects normal development and organogenesis in rice (Lu et al., 2011; Sun and Zhou, 2008). JMJ703 is implicated in the regulation of transposon activity (Cui et al., 2013). Besides, JmjC domain proteins can affect other epigenetic regulatory processes through their demethylation activities (Gan et al., 2014; Wu et al., 2019). Histone H3K9me2 demethylase can activate the expression of JMJ25 (Saze et al., 2008). The demethylase JMJ14 has H3K4me3/2/1 demethylation activity, and its mutants exhibit an early flowering phenotype (Searle et al., 2010).
In filamentous fungi, more and more protein complexes or chromatin proteins are also recruited to regulate chromatin functions, including gene expression, gene silencing, gene repair, and gene replication. In M. oryzea, until now, studies have shown that several mutants with altered conidium morphology, including Smo, MoIVD, Com1, MoSVP, and Con7, have been identified by chemical or insertional mutagenesis (Hamer et al., 1989; Li et al., 2004; Shi and Leung, 1995). MoIvd is highly conserved in fungi and the Δmoivd mutants showed reduced growth, decreased conidiation, and compromised pathogenicity, while the conidial germination and appressorial formation appeared normal (Li et al., 2019). Besides, Com1 encodes a putative transcription regulator unique to filamentous ascomycetes. The com1 disruption and deletion mutants had similar defects in conidium morphology and were significantly reduced in virulence on rice and barley seedlings (Yang et al., 2010). In addition, MoSVP can also act as a novel virulence effector which required for pathogenicity at the initial stage of infection (Shimizu et al., 2019). Con7 encodes a transcription factor with a zinc finger motif and a nuclear localization signal (Odenbach et al., 2007). In M. oryzae, our group previously purified the H3K4me3 methylation complex named COMPASS and proved its important roles in fungal development (Zhou et al., 2021). In 2023, we also reported that a cytoplasmic protein MoJMJD3 can reduce early appressorium formation and fungal virulence (Zhang et al., 2023a). The ΔMojmjd3 deletion mutants showed defects in early appressorium formation and fungal virulence during pathogenesis (Pham et al., 2015; Zhang et al., 2023a; Zhou et al., 2021). In addition, MoJMJ1 was been identified to encode an H3K4me3 histone demethylase containing JmjC domain which required for pathogenic development in M. oryzae. The MoJMJ1 regulated the expression of multiple target genes related to growth and development in M. oryzae (Huh et al., 2017).
Although proteins containing the JmjC domain exist widely in eukaryotes, the systematic properties of the JmjC domain family proteins, especially in terms of subcellular localization and biological function in M. oryzae, are rarely reported. In this study, we clearly revealed the JmjC domain-containing protein MoJMJD6 in M. oryzae, and then observed its subcellular localization and knockout mutant biological phenotype associated with fungal development and pathogenesis. GFP/RFP-MoJMJD6 is a fused protein localized in nuclei and fluorescence signals were involved in conidial germination and appressorium formation at the early stage of pathogenesis. The ΔMojmjd6 mutant strains showed defects in conidial germination and appressorium formation at the early stage of pathogenesis. This work will demonstrate the important roles of MoJMJD6 in fungal development and pathogenesis in M. oryzae.
Wild-type strain P131 of M. oryzae used in the experiment was incubated on oatmeal tomato agar medium (OTA) (75% oat juice [w/v], 15% tomato juice [w/v], and 2% agar powder [w/v]). The daily culture conditions for all strains were 26┬░C and light culture. The number of conidiation was measured as previously described (Zhou et al., 2021). Strains were cultured on oatmeal agar plates (40 g of oatmeal and 17 g of agar in 1,000 ml water) in the darkness at 25┬░C for 5 days. Then, for hyphal growth, small agar blocks were cut from the edge of 4-day-old cultures and placed into the complete medium for culture at 28┬░C with shaking at 180 rpm (Peng and Shishiyama, 1988). To measure the colony growth rate, mycelial of 5 mm were inoculated into the center of the OTA plates.
Nucleotide and protein sequences were obtained and analyzed using the rice blast database (http://fungi.ensembl.org/Magnaporthe_oryzae/Info/Index) and the National Center for Biotechnology Information (NCBI). Sequences of JmjC-domain-containing protein were obtained from the Rice Blast Database and ChromDB database (http://www.chromdb.org). Analyses using hidden Markov model profiling (http://hmmer.janelia.org) were performed to search for other JmjC proteins in M. oryzae and to analyze the conservation of the JmjC domain. A phylogenetic tree was constructed with MEGA 11 using the neighbor-joining method with a Poisson correction model and a bootstrap of 1,000 replicates.
For the preparation of M. oryzae protoplasts, the wild-type strain P131 was selected and grown on OTA plates for 4-6 days at 26┬░C. The hyphae were broken and transferred to 250 ml of complete medium for 36 h at 160 rpm and 28┬░C. Protoplasts were isolated using lysing enzymes Trichoderma harzianum-lyophilized powder (100 ╬╝g/ml) (L1412, Sigma, St. Louis, MO, USA). Collect protoplasts using 0.7 M NaCl buffer, 15 min at 4,000 rpm at 4┬░C. The precipitate was resuspended with STC solution (1.2 M D-sorbitol, 10 mM Tris-HCl pH 7.5, and 50 mM CaCl2). The protoplast concentration was adjusted to 1 ├Ś 108 protoplasts/ml with STC solution under a light microscope, and they were used for transformation.
For PEG-mediated transformation, add 150 ╬╝l of protoplasts and 2 ╬╝g of plasmids for 20 min at 4┬░C. Add 2 ml of PTC solution (60% PEG-3350, 1% Tris-HCl pH 7.5, and 0.05% CaCl2) for 20 min at 4┬░C and 30 ml of STC solution, 15 min at 4,000 rpm at 4┬░C. The supernatant was discarded, and 3 ml of LR medium (0.1% casein enzymatic hydrolysate, 0.1% yeast extract, and 34% sucrose) was added to the centrifuge tube. The tube was incubated in the light for 14-18 h at 28┬░C. The cultured protoplasts were added to a petri dish with a diameter of 10 cm, and 12 ml of SR (0.1% casein enzymatic hydrolysate, 0.1% yeast extract, 34% sucrose, and 0.7% agar) at a temperature of 50┬░C was added. After solidification, 20 ml of water agar medium containing antibiotics (500 mg/ml) was added. The plate was incubated in the light at 28┬░C for 4-6 days (Peng and Shishiyama, 1988).
For the localization analysis of MoJMJD6, the full length of MoJMJD6 was amplified using polymerase chain reaction (PCR) from wild-type P131 cDNA with primer MoJMJD6-GFP-F and MoJMJD6-GFP-R. The resulting fragment and p-rGTN vector were digested with EcoR I and Spe I, and MoJMJD6 was cloned into p-rGTN, which was generated by cloning the constitutive RP27 promoter into p-rGTN. The full length of MoJMJD6 was amplified using PCR from wild-type P131 of cDNA with primer MoJMJD6-RFP-F and MoJMJD6-RFP-R. The resulting fragment and p-rRTN vector were digested with EcoR I and Spe I, MoJMJD6 was cloned into p-rRTN. Protoplasts were isolated and transformed using PEG-mediated technology, refer to experimental method 2.3. The localization of the MoJMJD6 fusion proteins was determined with a Nikon A1 laser scanning confocal microscope (Tokyo, Japan). Detailed information on the primers used is shown in Table 1.
To construct gene deletion mutants, a homologous recombination strategy was used. The 1,500 bp upstream and downstream sequences of the gene MGG_01543 were extracted and named 1543-LB and 1543-RB. Combined with the multi-cloning site information on the pKOV21 vector, Spe I and EcoR I were selected as the endonucleases in the LB, and CIa I and Xho I were selected as the endonucleases for connecting in the RB. The 1543-LB and 1543-RB were amplified from wild-type P131 DNA using PCR with the primer pairs P1/P2 and P3/P4. The resulting fragments were linked into the pKOV21 vector containing hygromycin B phosphorylation transferase (HYG) (250 ╬╝g/ml).
First, in protoplast isolation and transformation experiment, the cell walls were dissolved with lysing enzymes to obtain protoplasts and linearized pKOV21 with Not I. After protoplast transformation, MoJMJD6 was replaced by hygromycin B through gene homologous recombination (Peng and Shishiyama, 1988). Transformants were obtained by screening for hygromycin resistance. Then the DNA of the strains was extracted. And then DNA was verified by PCR with primers LB-F1/UP-R1, Down-F2/RB-R2, HYG-F/HYG-R, and MoJMJD6-F/MoJMJD6-R (Table 1).
Genomic DNA was extracted using phenol-chloroform-isoamyl alcohol method (Peng and Shishiyama, 1988). Mycelia used for DNA extra were incubated in liquid complete medium (0.6% yeast extract, 0.3% enzymatic casein hydrolysate, 0.3% acidic casein hydrolysate, and 1% glucose). Two to three grams of mycelia were harvested and ground into a fine powder and then added CTAB at 65┬░C for 30 min. Then add 650 ╬╝l phenol-chloroform-isoamylol (25:24:1) 12,000 rpm for 10 min at 4┬░C. Take the supernatant and wash the precipitate with 1 ml 100% absolute ethanol and 1 ml 70% absolute ethanol, dissolved in 30 ╬╝l tris-EDTA buffer containing RNase to dissolve DNA. To prepare a 50 ╬╝l reaction system, add the following solutions to the PCR plate in sequence: DNA 2 ╬╝l, primer1 1 ╬╝l, primer2 2 ╬╝l, dNTPs 4 ╬╝l, 10├Ś buffer 5 ╬╝l, ddH2O 36.5 ╬╝l, and Taq enzyme 0.5 ╬╝l. DNA was pre-denatured at 95┬░C for 5 min, denatured at 95┬░C for 30 s, annealed at 58┬░C for 3 min, and extended at 72┬░C for 10 min, store at 4┬░C.
The conidia suspension of the test strains were prepared, and the concentration of the conidia suspension was adjusted to 5 ├Ś 104 conidia/ml using a hemocytometer under a microscope. The inner surface layer of fresh red onion was selected, cut into uniform squares (1 ├Ś 1 cm) with a sterile scalpel, and transferred to a water agar culture medium with sterile forceps. A pipette was used to evenly drop the conidial suspension on the inner and outer cells of the onion, and the prepared onions were incubated in a humidity box at 26┬░C. The infection process of Mojmjd6-RFP strain was analyzed using a Nikon A1 laser scanning confocal microscope.
Similarly, the conidial suspensions of the knockout mutant ΔMojmjd6 and the wild-type P131 were inoculated into onion epidermis. At 2 h, 4 h, and 8 h after inoculation, the germination of conidia was analyzed with a Nikon optical microscope. One hundred conidia were randomly selected to count the number of germinations. At 8 h, 12 h, and 24 h after inoculation, the formation of the appressorium was analyzed with a Nikon optical microscope. One hundred germinated conidia were randomly selected, and the number of appressoria formed was determined. Each experiment was repeated three times.
For pathogenicity tests and wound inoculation, M. oryzae conidia were collected from 7-day-old OTA plates and resuspended to 5 ├Ś 104 conidia/ml in 0.025% Tween-20 solution. Four-week-old seedlings of rice (Oryza sativa ŌĆśLTHŌĆÖ, rice varieties is Lijiang Xintuan Heigu [LTH]) were used for spray infection assays. The rice seedlings were grown in vermiculite supplied with liquid fertilizer in plastic pots (5 cm ├Ś 15 cm ├Ś 10 cm) at 25┬░C in a controlled-environment incubator with a 12-h photoperiod. The spore suspension was sprayed onto 4-week-old rice seedlings, which were then placed in a dark, humid dew chamber for 36 h at 25┬░C. Next, the seedlings were moved to an incubator at 25┬░C with a 12-h photoperiod for 5 days. Rice seedling leaves were collected on day 5 after the inoculation and analyzed. All inoculation and conidiation experiments were repeated three times independently.
For all tested strains, total proteins were extracted as described previously (Cao et al., 2016). One to two grams of mycelia was collected, and 1 ml of protein lysing buffer was added (50 mM Tris-HCl, pH 7.4, 150 mM NaCI, 1 mM EDTA, and 1% Triton X-100). Ten microliters of phenylmethanesulfonylfluoride and 10 ╬╝l protease inhibitor cocktail were added. After being mixed evenly, the prepared mycelia were then centrifuged for 20 min at 12,000 rpm at 4┬░C. The supernatant was collected and stored at ŌłÆ80┬░C.
The proteins of wild-type P131 and ΔMojmjd3 mutant were separated by 13% denaturing polyacrylamide gel (sodium dodecyl sulfate polyacrylamide gel electrophoresis), at 120 V for 1 h and transferred to the polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA) with a Bio-Rad electroblotting apparatus, at 150 mA for 1.2 h. The blots were probed with polyclonal histone H3 and histone methylation antibodies. First, the histones of wild-type P131 and knockout mutant ΔMojmjd6 were quantified with H3 antibody. Finally, the histone methylation-modified antibody was used for detection. The following antibodies were used: anti-H3 (ab1791, Abcam, Cambridge, UK). anti-H3K4me3 (ab8580, Abcam), anti-H3K9me3 (ab8898, Abcam), anti-H3K27me3 (ab6002, Abcam), and anti-H3K36me3 (ab9050, Abcam). The secondary antibody was HRP-labeled goat anti-rabbit IgG (H+L) antibody (HS101-01, Trans, Beijing, China).
The experimental data were analyzed by one-way ANOVA to determine the significance between treatments and expressed as P-value. All data are mean values for each treatment. Standard errors are calculated with the function STDEVP. When calculating percentages, the average number of conidia in each replicate was 100. Each test was repeated three times.
Phylogenetic analysis of JmjC-containing family members in M. oryzae. To explore whether the JmjC domain members was conserved in M. oryzae, a phylogenetic analysis of the JmjC-domain-containing homologues in the M. oryzae protein database was performed. Eleven putative demethylase genes with conserved JmjC domains were identified using blast analysis in the M. oryzae database (Fig. 1). These JmjC-domain-containing homologues were named MoJMJ1-MoJMJD1 1 (MoJMJ1, MGG_04878; MoJMJD2, MGG_09186; MoJMJD3, MGG_02032; MoJMJD4, MGG_04401; MoJMJD5, MGG_01068; MoJMJD6, MGG_01543; MoJMJD7, MGG_09841; MoJMJD8, MGG_02045; MoJMJD9, MGG_00445; MoJMJD10, MGG_03140; MoJMJD11, MGG_08136). MoJMJ1 (MGG_04878) was previously shown to be a histone H3K4me3 demethylase that is required for the pathogenic development of the rice blast fungus (Huh et al., 2017).
A phylogenetic analysis of these JmjC-domain-containing proteins was performed using MEGA11 software (Fig. 1). These 11 proteins both contain conserved functional domains such as JmjC, PHD, SET, F-box, and ARID, suggesting that these domains may play important roles in histone demethylation and methylation during fungal pathogenesis. Among other 10 proteins containing conserved JmjC domains, this study focused on MoJMJD6 (MGG_01543), which has a JmjC domain, for further analysis. The MoJMJD6 protein was predicted to have a JmjC domain at the C terminal of amino acids 257-532.
MoJMJD6 is localized in the nucleus of conidia, and participate in appressorium and invasive hyphae formation. In order to determine the location of MoJMJD6, both GFP-MoJMJD6 (green fluorescent protein [GFP]) and RFP-MoJMJD6 (red fluorescent protein [RFP]) fluorescent vector were constructed respectively and Fluorescence localization analysis were performed (Fig. 2A). The GFP:MoJMJD6 and RFP:MoJMJD6 fluorescence signals were both observed under a confocal laser-scanning microscope (LSM710, Zeiss) and MoJMJD6 was localized in the nucleus of conidia (Fig. 2B). As shown in Fig. 2C, MoJMJD6 participates in conidia germination, formation of appressorium and infective hyphae formation. These results showed that MoJMJD6 may have an important role in nucleus, mycelium, conidia, appressorium, and infectious hyphae of M. oryzae.
To determine whether Mojmjd6 was involved in the infection process of M. oryzae in the inner and outer cells of onion, infection experiments were performed on onion surface cells with the conidial suspension of the Mojmjd6-GFP and Mojmjd6-RFP strains. After inoculation, the RFP:MoJMJD6 signals of conidium germination stage (1 h), appressorium formation stage (6 h and 12 h), infection hyphae (24 h, 36 h, and 48 h) were observed (Fig. 2C). Indeed, we also observed the GFP:MoJMJD6 signals in conidia, hyphae, appressorium, and infectious hyphae during the pathogenesis. These results show that MoJMJD6 is involved in the infection process of M. oryzae, and may play important roles during pathogenesis.
The construction of a gene knockout vector strategy was used for the deletion of Mojmjd6 based on homologous recombination. The coding region of Mojmjd6 was replaced with HYG (hygromycin gene) (Fig. 3A). First, the LB and RB ofMoJMJD6 were amplified (Fig. 3B). The deletion mutants ╬öMojmjd6-1 and ╬öMojmjd6-2 were screened and confirmed using PCR (Fig. 3C). Furthermore, the hygromycin sequence (HYG) was amplified from the ╬öMojmjd6-1 and ╬öMojmjd6-2 (Fig. 3D). The colony size and the conidial formation of the mutants were compared with size of the wild-type on per plate (Fig. 4A). The colony diameter of ╬öMojmjd3 compared with the wild-type P131 strains on per plate after a 5-day incubation on OTA at 25┬░C, the colony diameter of P131 was 39.5 mm and ╬öMojmjd3 was 38.7 mm at the fifth day (Fig. 4B). The ╬öMojmjd6 mutants and wild-type P131 were grown on OTA plates for 8 days. The ╬öMojmjd6-1 mutant produce about 4.9 ┬▒ 0.15 ├Ś108 conidia and ╬öMojmjd6-2 mutant producing 4.95 ┬▒ 0.13 ├Ś 108 conidia per plate after an 8-day incubation on OTA at 25┬░C, compared with the wild-type P131 which produced 5.2 ┬▒ 0.21 ├Ś 108 conidia per plate after an 8-day incubation on OTA at 25 ┬░C (P > 0.05) (Fig. 4C). These data showed that there were no significance differences between the wild-type and the ╬öMojmjd6-1 and ╬öMojmjd6-2 deletion mutants and there was no significant difference in colony diameter and cnidogenesis.
To further determine whether deletion of Mojmjd6 affects the pathogenesis, inoculating the conidial suspensions of the wild-type strain and the ΔMojmjd6 mutants onto onion epidermal cells. Interestingly, the conidia germination of ΔMojmjd6 was significantly decreased when compared with wild-type P131 at 2 dpi and 4 dpi (P = 0.03) (Fig. 5A and B). In addition, the appressorium growth of ΔMojmjd6 was significantly decreased when compared with wild-type P131 at 4 dpi and 8 dpi (P = 0.02) (Fig. 5C). These results show that Mojmjd6 may play important roles in the early stage of conidial germination rate and appressorium formation during the early stage of pathogenesis.
Since we observed there are some defects of the Mojmjd6 deletion mutants during the conidial germination and appressorium formation, we next investigated whether the glycogen and lipid, some conidial storage, was blocked. We then used I2/KI solution and Nile Red to stain the glycogen, and lipid during appressorium formation, respectively. As shown in Fig. 5, in the wild type, along with conidium germination and appressorium formation, glycogen was transferred from conidia to the appressorium at 8 dpi, while in the ΔMojmjd6 deletion mutants, glycogen could be observed until 12 dpi and has defects in utilization at 24 dpi (Fig. 5D). Constantly, the wild type P131 has transferred lipid from the conidium to appressorium at 12 dpi, while the ΔMojmjd6 mutant has defects in lipid utilization until 12 dpi (Fig. 5E). These results suggested that the glycogen utilization and lipid transformation was blocked in the ΔMojmjd6 mutant strains.
To further investigate whether Mojmjd6 plays an important role in the pathogenic mechanism of M. oryzae, virulence tests were conducted on rice seedlings with mycelial blocks and conidial spray inoculation, and the symptom development at 5 dpi was compared. These results showed a virulence reduction of the ΔMojmjd6 deletion mutants on the leaves of rice seedlings (Fig. 6A and B). The typical spindle-shaped lesions were observed on the leaves challenged with the wild-type strain, while leaves inoculated with the deletion mutants exhibited smaller lesions (Fig. 6A and B). In addition, we also counted the area of pathogenic lesions on rice by wild type and mutant mycelium block inoculation and the number of lesions produced by conidial suspension inoculation on rice (Fig. 6C and D, Supplementary Tables 1 and 2). According to statistical analysis, the pathogenic area of ΔMojmjd6 strain was reduced compared with the wild type, and the number of disease spots was also decreased. These results suggested that deletion of MoJMJD6 can reduce the virulence of M. oryzae to rice.
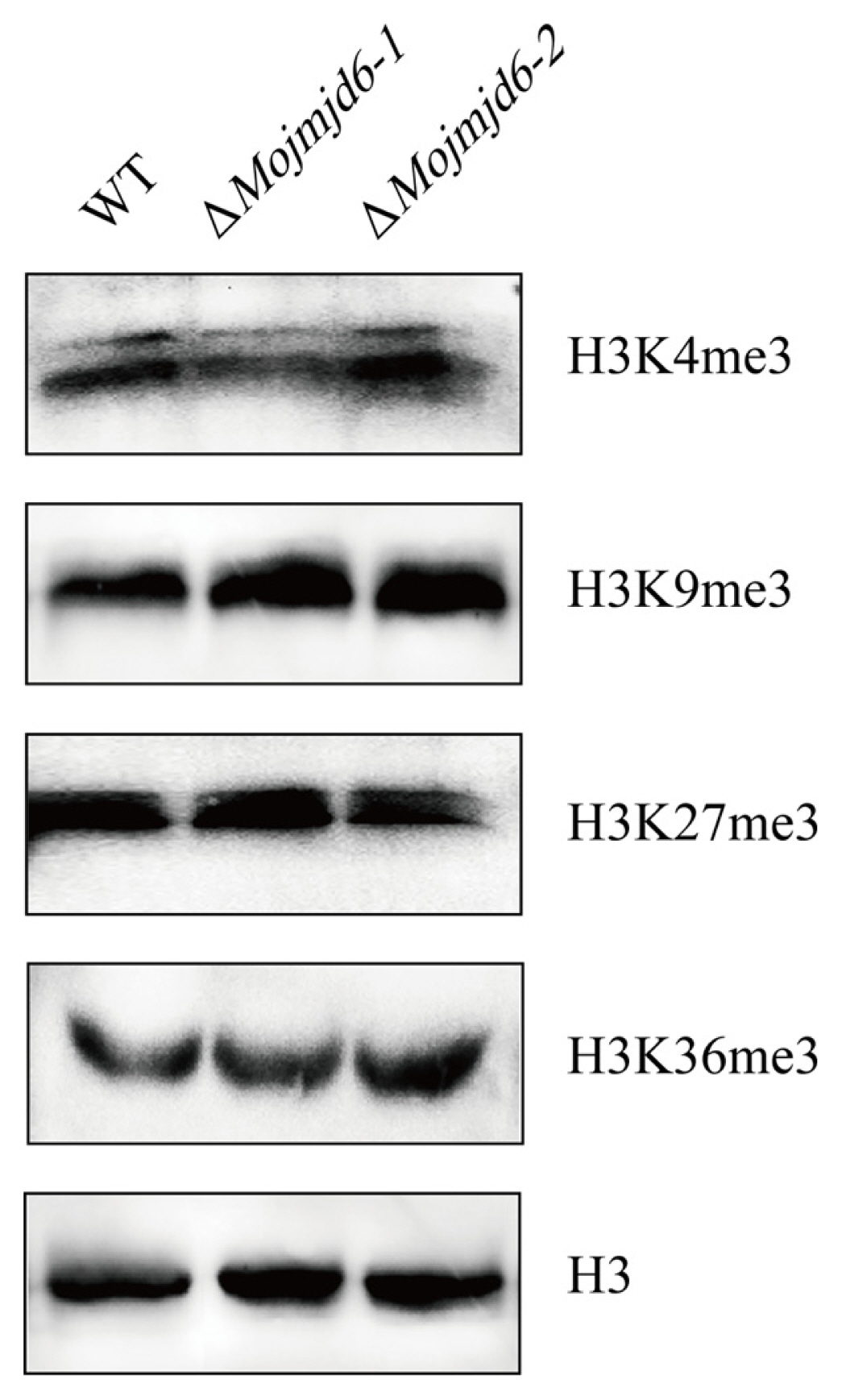
In order to investigate the function of a JmjC group demethylase in fungal development after the Mojmjd6 gene was knocked out. We performed a western blot analysis to detect whether there were changes in the genome-wide histone methylation levels in M. oryzae using the antibodies raised against H3K4me3, H3K9me3, H3K36me3, or H3K27me3. Total proteins were extracted from the mycelia of ΔMojmjd6 mutant and wild-type strains P131. But there was no significant change in the signal of H3K4me3, H3K9me3, H3K27me3, and H3K36me3 methylation in the ΔMojmjd6 mutant when compared with the wild-type strain P131 (Fig. 7). These results showed that MoJMJD6 may have no histone demethylase activity at the genome-wide chromatin, which suggesting this MoJMJD6 perhaps is a more specific histone modifier for regulating some key genes during pathogenesis.
The JmjC domain, which mediates lysine methylation, is one of the major epigenetic marks and regulates chromatin-mediated gene transcription. At present, a large number of studies have been carried out to analyze the pathogenic mechanism of the rice blast. Many pathogenic genes are involved in the attachment and germination of conidia, the formation and maturation of appressoria, the generation of infection spikes, and the differentiation and expansion of infected hyphae. Previous studies have shown that MoSET1, plays an important role in regulating genes during morphogenesis (Pham et al., 2015). MoPmk1 affects appressorium formation in M. oryzae and controls the expansion of hyphae to neighboring cells (Sakulkoo et al., 2018). MoMst50 is also critical for appressorium formation (Park et al., 2006). The ΔMofap7 mutant can affect the growth and infection of M. oryzae (Li et al., 2017). And the ΔMorac1 mutant was found to slower growth rate, conidial shape changes, and reduced pathogenicity of M. oryzae (Chen et al., 2008). In 2023, we identified a RNAPII degradation factor Def1 which is required for development, stress response, and full virulence of M. oryzae (Zhang et al., 2023b).
Epigenetic mechanisms in host-pathogen interactions lay the foundation for future fungal research. Understanding the process by which pathogens infect plants plays an important role in reducing pathogen damage to the host which may help plants fight disease. As we know that, histone demethylation modification is an important way of histone modification in chromatin modification. Lysine-specific demethylase 1 and JmjC domain-containing proteins as two histone demethylases have been reported to be involved in plant vegetative growth, reproductive development, and disease resistance. In 2021, our research group purified the COMPASS complex with Flag-tag affinity, and demonstrated that the deletion of the key subunits encoding MoBre2, MoSPP1, and MoSwd2 resulted in defects during the development of invasive hyphae in M. oryzae (Zhou et al., 2021). During rice cell colonization, M. oryzae must suppress or avoid triggering two types of innate immunity, pathogen-associated molecular pattern-triggered immunity, and effector-triggered immunity (Barski et al., 2007).
In recent years, many functional genes such as MoRPD3, MoUbp8, Pal1, MoEmc2, PoRal2, and so on, were both involved in appressoria formation, conidiation, hyphae growth (Chen et al., 2022; Lee et al., 2021; Qu et al., 2021; Yang et al., 2020; Yu et al., 2021). This study preliminarily explores MoJMJD6 in the growth, development, and pathogenic mechanism of M. oryzae, which may provide new ideas for the development of fungal disease control strategies and new candidate targets for the development of new fungicides in plant fungi. However, there is a subtle epigenetic mechanism during appressoria repolarization to organize the penetration to host cells, but whether the histone demethylation defect in penetration of ΔMojmjd6 mutant has resulted from the defective appressoria development needs more evidence. Future study will discover how epigenetic histone methylations and histone demethylations cross talk and bivalent regulate key effectors such as DNA double-strand break repair, cell cycle regulation or virulence in fungi and inflammation modulation in human and animals which will be a novel player in potential biomarker for fungicides and human tumors.
We concluded that MoJMJD6 protein containing a JmjC domain in the rice blast fungus is located in the nucleus, and the deletion of the Mojmjd6 is required for conidiation, conidium germination, appressorium formation, and defective in glycogen and lipid utilization during pathogenicity on rice cultivar. In this study, we generated ΔMojmjd6 strains from the P131 wild-type strain, the ΔMojmjd6 knockout mutants showed defects in conidial germination and appressorium formation at 2 dpi, 4 dpi, and 8 dpi post inoculation which suggests that Mojmjd6 may function in the early stage of pathogenesis.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grants 31871638 to W.W.), the Special Scientific Research Project of Beijing Agriculture University (YQ201603), the Research Fund for Academic Degree & Graduate Education of Beijing University of Agriculture (2019YJS037), the Research Fund of State Key Laboratory for Biology of Plant Diseases and Insect Pests (SKLOF202102).
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig.┬Ā1
MoJMJD6 is a conserved JmjC domain-containing protein. Distribution and protein domain description of demethylase genes contain the JmjC domain in Magnaporthe oryzae. The phylogenetic tree of JmjC demethylase gene in rice blast fungus was constructed in MEGA11 software using the neighbor-joining method with a Poisson correction model and a bootstrap of 1,000 replicates. Information about domain architectures was obtained from InterProScan. Details of all analyzed gene sequences and amino acid sequences are shown in Supplementary Fig. 1.
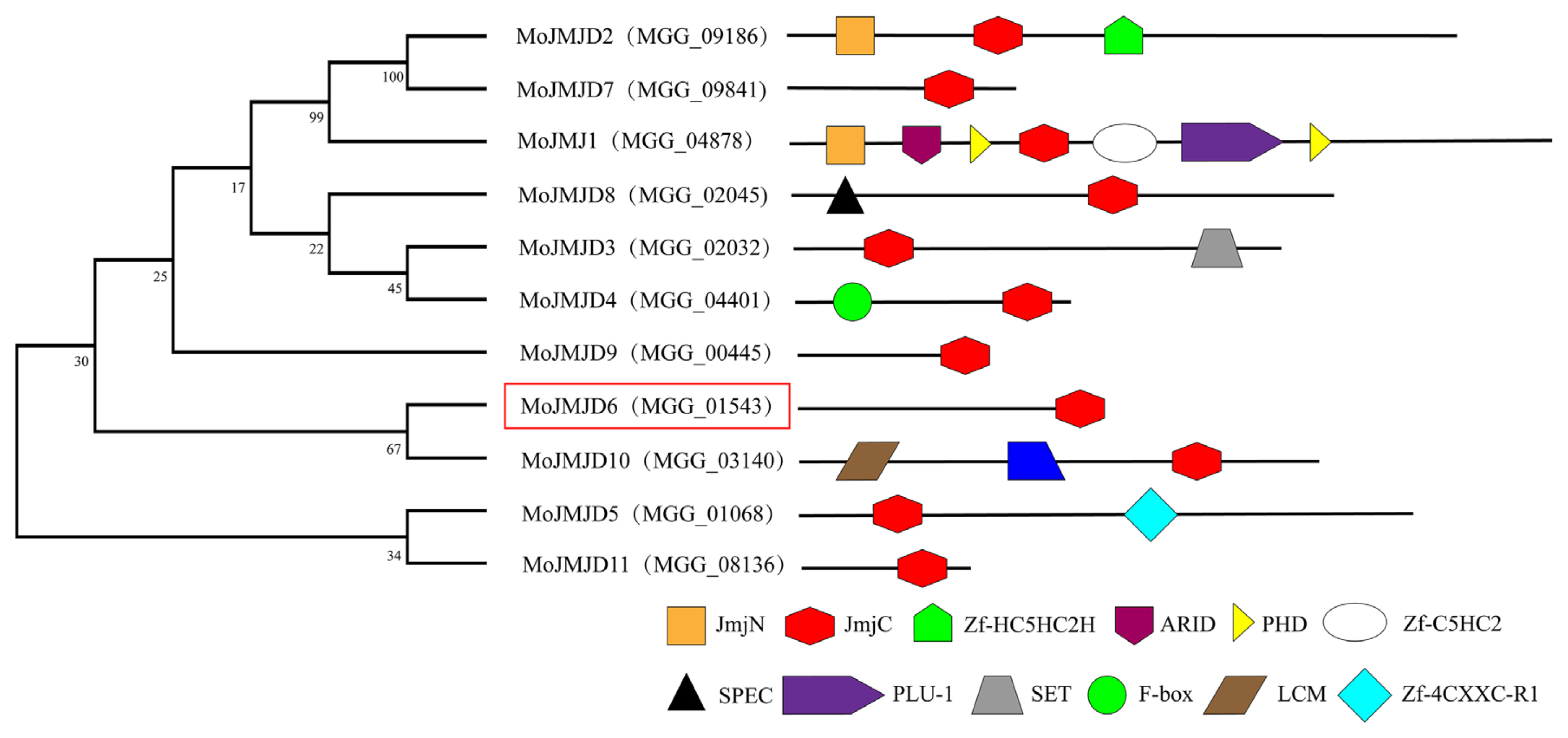
Fig.┬Ā2
Fluorescence subcellular localization analysis of MoJMJD6. (A) Schematic diagram of the construction of the Mojmjd6-pGTN/pRTN eukaryotic fluorescent fusion vector. The full-length Mojmjd6 was ligated into the eukaryotic fluorescent expression vector. Fluorescence subcellular localization of MoJMJD6-GFP in the nucleus of Magnaporthe oryzae. (B) Fluorescence subcellular localization of MoJMJD6, localized in the nucleus of M. oryzae. Scale bars = 250 ╬╝m. (C) Analysis of the infection process of the fluorescent strain Mojmjd6-RFP inoculated on the inner and outer layers of onion. After inoculation, we focus on the conidium germination stage (1-6 h), appressorium formation stage (12-24 h), infection spike production stage (36 h), and infective hyphae formation stage (48 h) of M. oryzae. Scale bars = 50 ╬╝m.
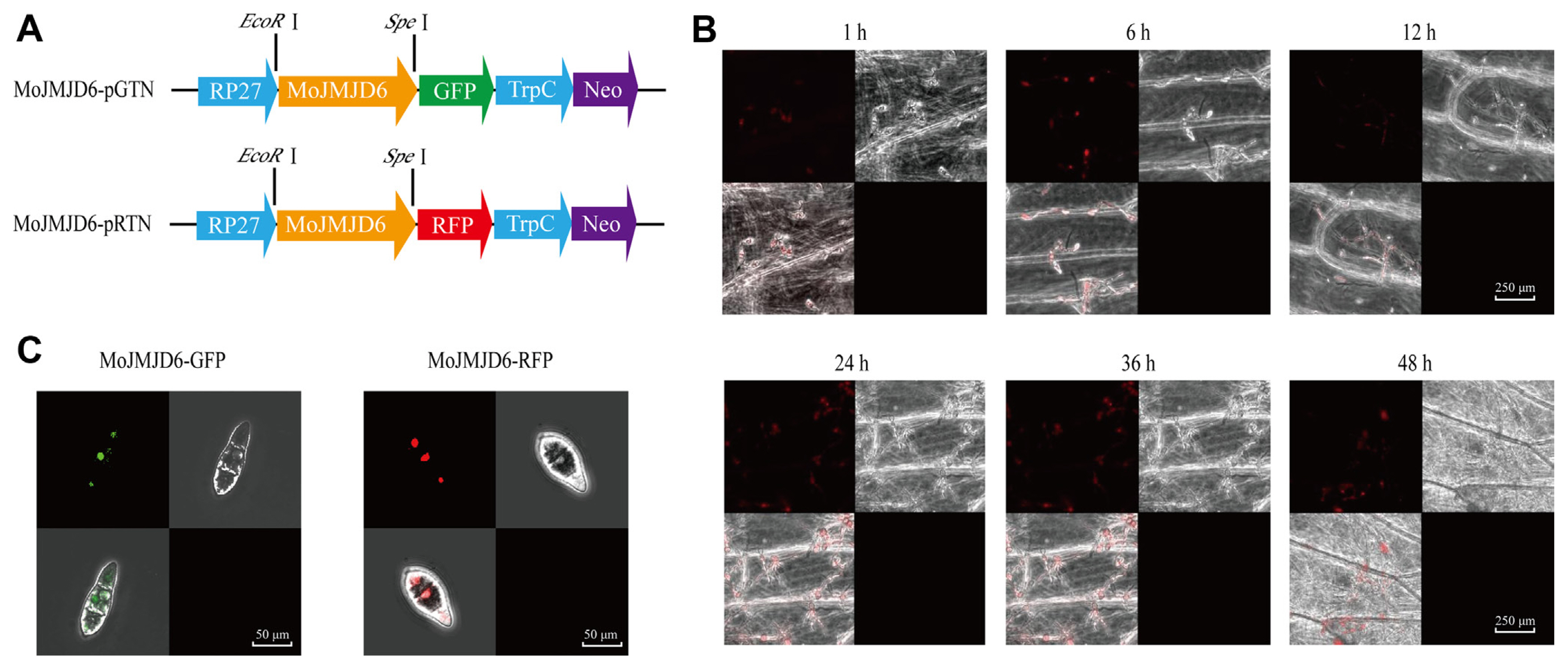
Fig.┬Ā3
Construction and acquisition of the knockout mutant strain ΔMojmjd6. (A) Schematic diagram of targeted gene replacement of Mojmjd6 in Magnaporthe oryzae. (B) The polymerase chain reaction (PCR) amplification of 1543-LB and 1543-RB. The flanking sequences (1543-LB and 1543-RB) of Mojmjd6 were amplified using PCR in vitro with the wild-type P131 genomic DNA of M. oryzae. (C) The PCR amplification results of the Mojmjd6 gene. The Mojmjd6 gene was amplified using PCR with the genomic DNA of the knockout mutant ΔMojmjd6 and the wild-type P131 as templates. (D) PCR verification results using ΔMojmjd6 knockout DNA as a template. The hygromycin sequence (HYG) was identified from the genomic DNA template of the knockout mutant ΔMojmjd6, as were the sequences between LB and RB of Mojmjd6 to hygromycin (LB-HY and RB-YG).
Fig.┬Ā4
Phenotypic characterization of ΔMojmjd6 mutants. (A) Colony morphology of the knockout mutant ΔMojmjd6 and the wild-type P131 grown on oatmeal tomato agar medium for 7 days. (B) The growth rate of the knockout ΔMojmjd6 versus the wild-type P131 before 5 days. Three independent experiments were repeated and the average was taken for analysis. (C) Analysis of the sporulation yield of the knockout mutant ΔMojmjd6 and the wild-type P131. Three independent experiments were repeated and the average was taken for analysis.

Fig.┬Ā5
Phenotypic differences of the knockout mutant ΔMojmjd6 and the wild-type P131 during rice blast fungus infection. (A) The morphological characteristics of the knockout mutant ΔMojmjd6 and the wild-type P131 in different time periods of onion infection. Scale bar = 20 μm. (B) Comparison of the knockout mutant ΔMojmjd6 and the wild-type P131 at the conidial germination stage of Magnaporthe oryzae. One hundred conidia were randomly selected, the number of conidia germinated was counted, three independent experiments were performed, and average value was taken for analysis (*P < 0.05). (C) Comparison of the knockout mutant ΔMojmjd6 and the wild-type P131 in the appressorium formation stage of M. oryzae. One hundred germinated conidia were randomly selected, the number of appressoria formed was counted, the independent experiments was repeated three times, and the average value was taken for analysis (*P < 0.05). (D, E) MoJMJD6 affects utilization of glycogen and lipid transformation during appressorium development. I2/KI solution (D) and Nile red (E) staining of the wild type and the ΔMojmjd6 mutants. Glycogen was stained with yellowish brown color and lipid was stained and fluoresce red. Scale bars = 20 μm.
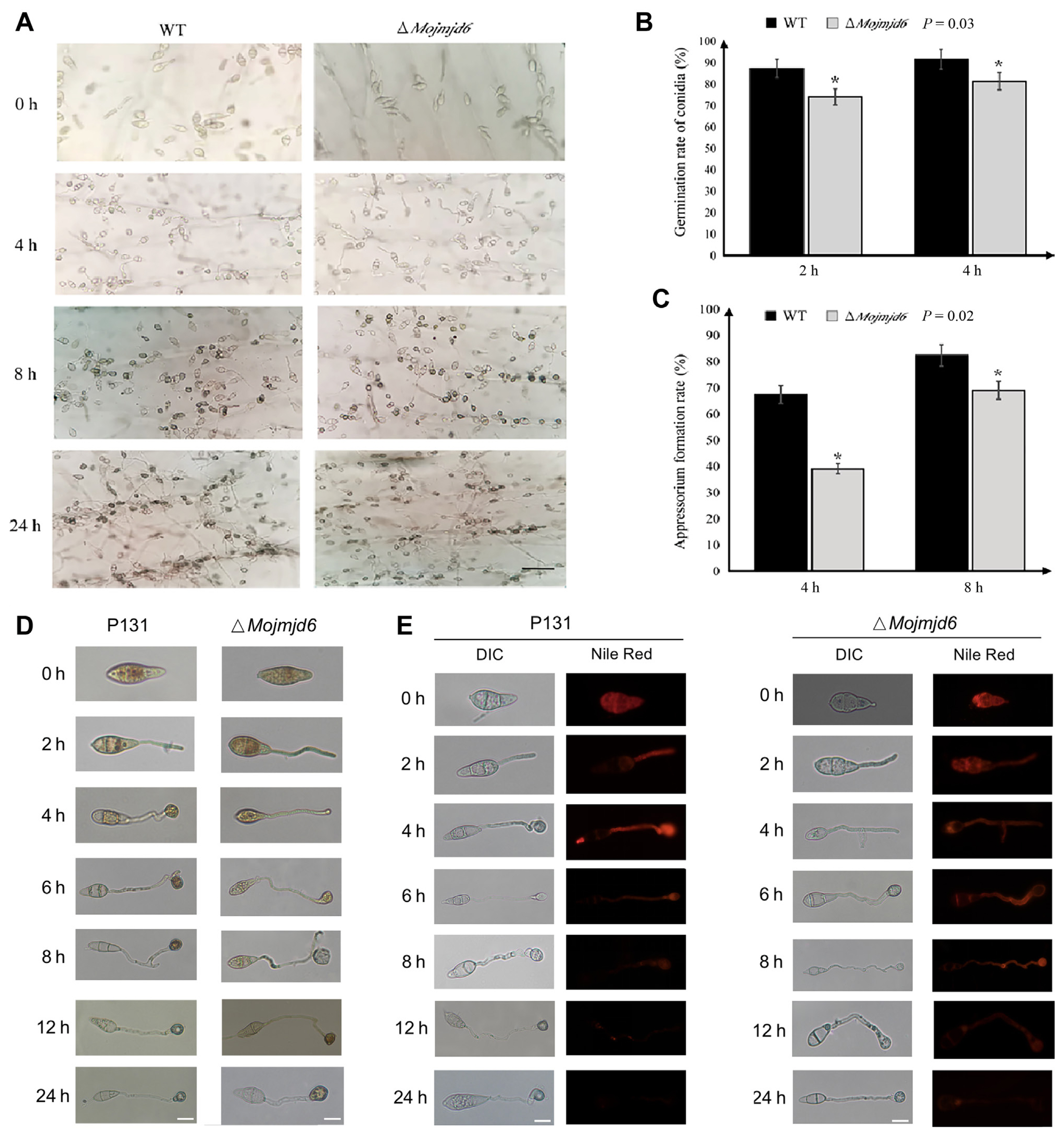
Fig.┬Ā6
Pathogenicity identification of the knockout mutant ΔMojmjd6. (A) The mycelial blocks of the knockout mutant ΔMojmjd6 and the wild-type P131 were inoculated on scratched rice leaves. Three mycelial blocks were connected to each rice leaf, and three independent experiments were repeated. (B) Disease incidence of the knockout mutant ΔMojmjd6 and the wild-type P131 conidial suspensions sprayed on rice leaves. Three independent experiments were repeated. (C) The area of lesions on each leaf (mm), each test was repeated three times as shown in the histogram. (D) Number of lesions per leaf, each test was repeated three times as shown in the histogram.
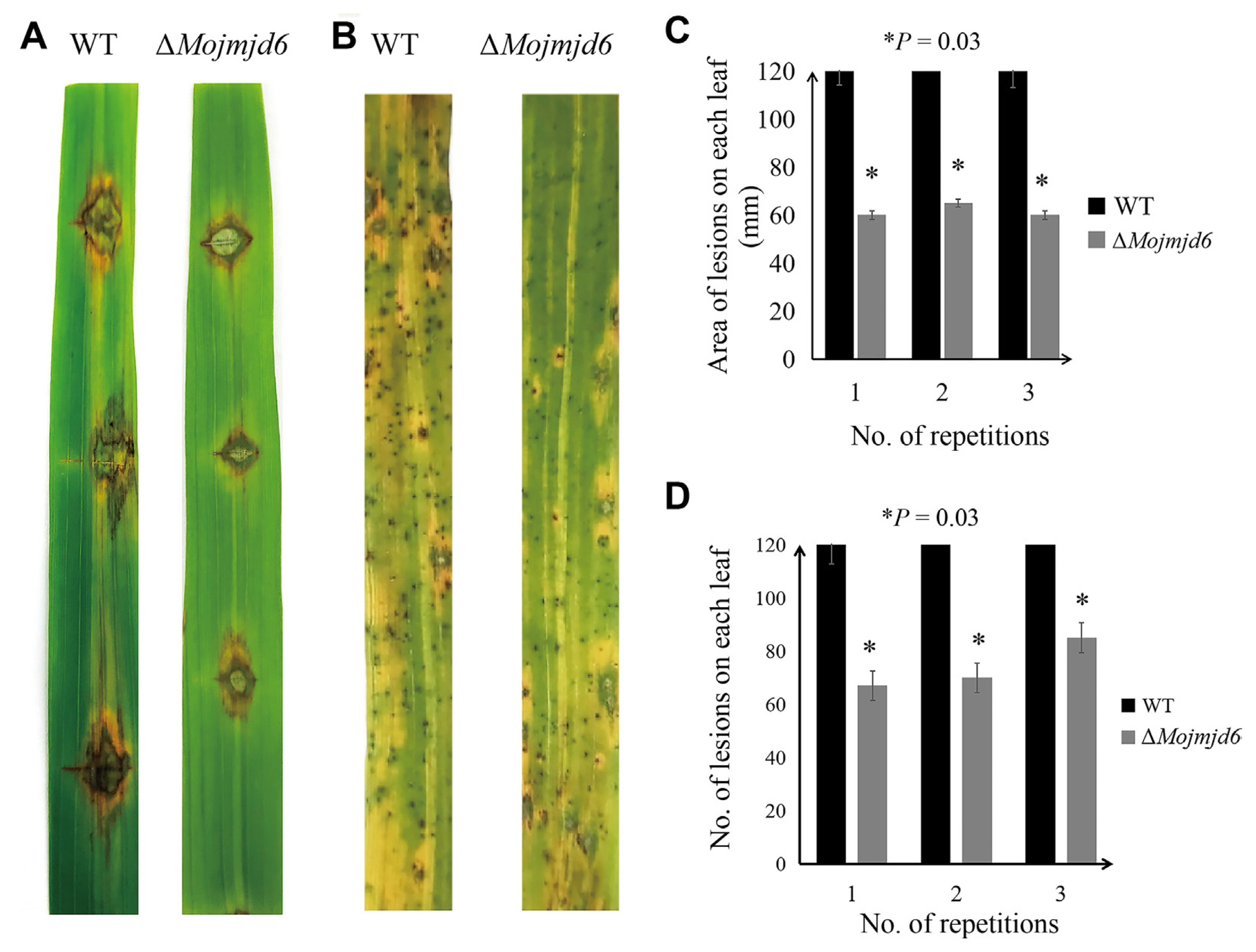
Fig.┬Ā7
Detection of histone methylation levels in Magnaporthe oryzae. The total histones were extracted from mycelia of wild-type and ΔMojmjd6 mutants and separated on a 13% sodium dodecyl sulfate polyacrylamide gel electrophoresis gel and transferred to a polyvinylidene difluoride membrane. Blot was probed with polyclonal anti-H3 antibody and Histone methylation antibodies (H3K4me3, H3K9me3, H3K27me3, and H3K36me3). H3-antibody was used to quantify the histone protein of the wild-type and knockout mutants ΔMojmjd6.
Table┬Ā1
Primer sequence were used in this study
References
Agger, K., Cloos, P. A. C., Rudkj├”r, L., Williams, K., Andersen, G., Christensen, J. and Helin, K. 2009. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev 23:1171-1176.



Barski, A., Cuddapah, S., Cui, K., Roh, T.-Y., Schones, D. E., Wang, Z., Wei, G., Chepelev, I. and Zhao, K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823-837.


Cao, Z., Yin, Y., Sun, X., Han, J., Sun, Q. P., Lu, M., Pan, J. and Wang, W. 2016. An ash1-like protein MoKMT2H null mutant is delayed for conidium germination and pathogenesis in Magnaporthe oryzae. Biomed Res. Int 2016:1575430.




Chen, D., Hu, H., He, W., Zhang, S., Tang, M., Xiang, S., Liu, C., Cai, X., Hendy, A., Kamran, M., Liu, H., Zheng, L., Huang, J., Chen, X.-L. and Xing, J. 2022. Endocytic protein Pal1 regulates appressorium formation and is required for full virulence of Magnaporthe oryzae. Mol. Plant Pathol 23:133-147.



Chen, J., Zheng, W., Zheng, S., Zhang, D., Sang, W., Chen, X., Li, G., Lu, G. and Wang, Z. 2008. Rac1 is required for pathogenicity and Chm1-dependent conidiogenesis in rice fungal pathogen Magnaporthe grisea. PLoS Pathog 4:e1000202.



Cui, X., Jin, P., Cui, X., Gu, L., Lu, Z., Xue, Y., Wei, L., Qi, J., Song, X., Luo, M., An, G. and Cao, X. 2013. Control of transposon activity by a histone H3K4 demethylase in rice. Proc. Natl. Acad. Sci. U. S. A 110:1953-1958.



Gan, E.-S., Xu, Y., Wong, J.-Y., Goh, J. G., Sun, B., Wee, W.-Y., Huang, J. and Ito, T. 2014. Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat. Commun 5:5098.



Hamer, J. E., Valent, B. and Chumley, F. G. 1989. Mutations at the smo genetic locus affect the shape of diverse cell types in the rice blast fungus. Genetics 122:351-361.




Huang, S., Zhang, A., Jin, J. B., Zhao, B., Wang, T.-J., Wu, Y., Wang, S., Liu, Y., Wang, J., Guo, P., Ahmad, R., Liu, B. and Xu, Z.-Y. 2019. Arabidopsis histone H3K4 demethylase JMJ17 functions in dehydration stress response. New Phytol 223:1372-1387.

Huang, Y., Chen, D., Liu, C., Shen, W. and Ruan, Y. 2016. Evolution and conservation of JmjC domain proteins in the green lineage. Mol. Genet. Genomics 291:33-49.



Huh, A., Dubey, A., Kim, S., Jeon, J. and Lee, Y.-H. 2017. MoJMJ1, encoding a histone demethylase containing JmjC domain, is required for pathogenic development of the rice blast fungus, Magnaporthe oryzae. Plant Pathol. J 33:193-205.



Kankanala, P., Czymmek, K. and Valent, B. 2007. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19:706-724.




Klose, R. J., Kallin, E. M. and Zhang, Y. 2006. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet 7:715-727.



Kooistra, S. M. and Helin, K. 2012. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol 13:297-311.



Lee, S. H., Farh, ME-A, Lee, J., Oh, Y. T., Cho, E., Park, J., Son, H. and Jeon, J. 2021. A histone deacetylase, Magnaporthe oryzae RPD3, regulates reproduction and pathogenic development in the rice blast fungus. mBio 12:e0260021.



Li, G., Zhang, X., Tian, H., Choi, Y.-E., Tao, W. A. and Xu, J.-R. 2017. MST50 is involved in multiple MAP kinase signaling pathways in Magnaporthe oryzae. Environ. Microbiol 19:1959-1974.



Li, L., Xue, C., Bruno, K., Nishimura, M. and Xu, J.-R. 2004. Two PAK kinase genes, CHM1 and MST20, have distinct functions in Magnaporthe grisea. Mol. Plant-Microbe Interact 17:547-556.


Li, Y., Zheng, X., Zhu, M., Chen, M., Zhang, S., He, F., Chen, X., Lv, J., Pei, M., Zhang, Y., Zhang, Y., Wang, W., Zhang, J., Wang, M., Wang, Z., Li, G. and Lu, G. 2019. MoIVD-mediated leucine catabolism is required for vegetative growth, conidiation and full virulence of the rice blast fungus Magnaporthe oryzae. Front. Microbiol 10:444.



Liu, P., Zhang, S., Zhou, B., Luo, X., Zhou, X. F., Cai, B., Jin, Y. H., Niu, D., Lin, J., Cao, X. and Jin, J. B. 2019. The histone H3K4 demethylase JMJ16 represses leaf senescence in Arabidopsis. Plant Cell 31:430-443.



Lloret-Llinares, M., Carr├®, C., Vaquero, A., de Olano, N. and Azor├Łn, F. 2008. Characterization of Drosophila melanogaster JmjC+N histone demethylases. Nucleic Acids Res 36:2852-2863.



Lu, F., Cui, X., Zhang, S., Jenuwein, T. and Cao, X. 2011. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet 43:715-719.



Mosammaparast, N. and Shi, Y. 2010. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem 79:155-179.


Odenbach, D., Breth, B., Thines, E., Weber, R. W. S., Anke, H. and Foster, A. J. 2007. The transcription factor Con7p is a central regulator of infection-related morphogenesis in the rice blast fungus Magnaporthe grisea. Mol. Microbiol 64:293-307.


Park, G., Xue, C., Zhao, X., Kim, Y., Orbach, M. and Xu, J.-R. 2006. Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea. Plant Cell 18:2822-2835.




Peng, Y.-L. and Shishiyama, J. 1988. Temporal sequence of cytological events in rice leaves infected with Pyricularia oryzae. Can. J. Bot 66:730-735.

Pham, K. T. M., Inoue, Y., Vu, B. V., Nguyen, H. H., Nakayashiki, T., Ikeda, K.-I. and Nakayashiki, H. 2015. MoSET1 (histone H3K4 methyltransferase in Magnapothe oryzae) regulates global gene expression during infection-related morphogenesis. PLoS Genet 11:e1005385.



Pinnschmidt, H. O., Bonman, J. M. and Kranz, J. 1995. Lesion development and sporulation of rice blast. J. Plant Dis. Prot 102:299-306.
Qu, Y., Wang, J., Huang, P., Liu, X., Lu, J. and Lin, F.-C. 2021. PoRal2 is involved in appressorium formation and virulence via Pmk1 MAPK pathways in the rice blast fungus Pyricularia oryzae. Front. Plant Sci 12:702368.



Sakulkoo, W., Os├®s-Ruiz, M., Oliveira Garcia, E., Soanes, D. M., Littlejohn, G. R., Hacker, C., Correia, A., Valent, B. and Talbot, N. J. 2018. A single fungal MAP kinase controls plant cell-to-cell invasion by the rice blast fungus. Science 359:1399-1403.


Saze, H., Shiraishi, A., Miura, A. and Kakutani, T. 2008. Control of genic DNA methylation by a JmjC domain-containing protein in Arabidopsis thaliana. Science 319:462-465.


Searle, I. R., Pontes, O., Melnyk, C. W., Smith, L. M. and Baulcombe, D. C. 2010. JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev 24:986-991.



Shi, Z. and Leung, H. 1995. Genetic analysis of sporulation in Magnaporthe grisea by chemical and insertional mutagenesis. Mol. Plant-Microbe Interact 8:949-959.

Shimizu, M., Nakano, Y., Hirabuchi, A., Yoshino, K., Kobayashi, M., Yamamoto, K., Terauchi, R. and Saitoh, H. 2019. RNA-Seq of in planta-expressed Magnaporthe oryzae genes identifies MoSVP as a highly expressed gene required for pathogenicity at the initial stage of infection. Mol. Plant Pathol 20:1682-1695.




Sun, Q. and Zhou, D.-X. 2008. Rice JmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc. Natl. Acad. Sci. U. S. A 105:13679-13684.



Valent, B., Farrall, L. and Chumley, F. G. 1991. Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics 127:87-101.




Wu, J., Yamaguchi, N. and Ito, T. 2019. Histone demethylases control root elongation in response to stress-signaling hormone abscisic acid. Plant Signal. Behav 14:1604019.



Xu, J. R., Zhao, X. and Dean, R. A. 2007. From genes to genomes: a new paradigm for studying fungal pathogenesis in Magnaporthe oryzae. Adv. Genet 57:175-218.


Yamane, K., Toumazou, C., Tsukada, Y.-I., Erdjument-Bromage, H., Tempst, P., Wong, J. and Zhang, Y. 2006. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 5:483-495.

Yang, J., Chen, D., Matar, K. A. O., Zheng, T., Zhao, Q., Xie, Y., Gao, X., Li, M., Wang, B. and Lu, G.-D. 2020. The deubiquitinating enzyme MoUbp8 is required for infection-related development, pathogenicity, and carbon catabolite repression in Magnaporthe oryzae. Appl. Microbiol. Biotechnol 104:5081-5094.



Yang, J., Zhao, X., Sun, J., Kang, Z., Ding, S., Xu, J.-R. and Peng, Y.-L. 2010. A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae. Mol. Plant-Microbe Interact 23:112-123.


Yu, R., Shen, X., Liu, M., Liu, X., Yin, Z., Li, X., Feng, W., Hu, J., Zhang, H., Zheng, X., Wang, P. and Zhang, Z. 2021. The rice blast fungus MoRgs1 functioning in cAMP signaling and pathogenicity is regulated by casein kinase MoCk2 phosphorylation and modulated by membrane protein MoEmc2. PLoS Pathog 17:e1009657.



Zhang, L., Zhang, M., Liu, C., Wu, Z., Luo, Z., Lu, M., Shi, Y., Nan, Z., Hu, D., Pan, J. and Wang, W. 2023a. A cytoplasmic protein MoJMJD3 can reduce early appressorium formation and fungal virulence in Magnaporthe oryzae. J. Phytopathol 171:223-233.

- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 1,759 View
- 78 Download
- ORCID iDs
-
Weixiang Wang

https://orcid.org/0000-0001-8586-1499 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement1
Supplement1 Print
Print



