Generation and Retention of Defective RNA3 from Cucumber Mosaic Virus and Relevance of the 2b Protein to Generation of the Subviral RNA
Article information
Abstract
A defective RNA3 (D3Yα) of strain Y of cucumber mosaic virus (CMV-Y) was examined on host-specific maintenance, experimental conditions, and a viral factor required for its generation in plants. D3Yα was stably maintained in cucumber but not in tomato plants for 28 days post inoculation (dpi). D3Yα was generated in Nicotiana tabacum or N. benthamiana after prolonged infection in the second and the third passages, but not in plants of N. benthamiana grown at low temperature at 28 dpi or infected with CMV-Y mutant that had the 2b gene deleted. Collectively, we suggest that generation and retention of D3Yα depends on potential host plants and experimental conditions, and that the 2b protein has a role for facilitation of generation of D3Yα.
The genome of cucumber mosaic virus (CMV), a member of the genus Cucumovirus in the family Bromoviridae consists of three RNA segments designated as RNA1, RNA2, and RNA3 in descending order in length (Palukaitis and García-Arenal, 2003). RNA1 and RNA2 encode, respectively, the 1a and the 2a proteins, which are responsible for virus replication. RNA2 also contains a small open reading frame (ORF) for the 2b protein which is one of the best-studied RNA silencing suppressors of plant viruses (Nakahara and Masuta, 2014). RNA3 has two ORFs encoding the 3a protein and the coat protein (CP) that is required for viral cell-to-cell movement and encapsidation, respectively.
A defective (D) RNA of a plant virus can generate spontaneously during replication of the viral genome in the host cell (Graves et al., 1996; Hasiów-Jaroszewska et al., 2018; Simon et al., 2004). The D RNA is defective for independent replication and requires the missing functions provided by the complete virus genome (helper virus) (Takeshita et al., 2009). Two subviral RNAs associated with the Fny strain of CMV have been characterized as 3α-type and 3β-type D RNA3 and differ in the size of deletion in the 3a gene. According to Kaplan et al. (2004), a β-type D RNA3 from CMV-Fny (D RNA 3-1) is detected in tobacco but not in squash.
We previously determined the nucleotide sequence of an α-type D RNA3 from Y strain of CMV (CMV-Y) RNA3, named D RNA3Yα (D3Yα), and constructed an infective cDNA clone of D3Yα which was maintained stably by an original helper virus, CMV-Y, without any difference in symptoms from the original virus strain (Takeshita et al., 2008). Subsequently, we elucidated that spatial interference of D3Yα in accumulation patterns of RNA3 resulted in the indistinguishable symptoms of D3Yα from the original ones induced by the helper virus (Takeshita et al., 2009). While D3Yα can arise during CMV infection in Nicotiana tabacum cv. Samsun NN and N. benthamiana, its host-adaptability and the environmental and viral factors associated with its generation and accumulation have not been well characterized. In this study, we investigated host-specific maintenance of D3Yα and monitored generation of D3Yα using the infective cDNA clones of CMV-Y RNAs. In addition, we examined involvement of the 2b protein of CMV-Y in the generation and/or accumulation of D3Yα.
Materials and Methods
Plants and inoculation assays
Plants of N. tabacum cv. Samsun NN, N. benthamiana, cucumber, and tomato were grown in an air-conditioned greenhouse under natural light at 25°C/20°C (day/night). Inocula of CMV-Y virions and in vitro transcripts of D3Yα were prepared as reported by Takeshita et al. (2008). Absence of D3Yα in the purified CMV-Y virions was certified using 3a-ORF-specific reverse transcription polymerase chain reaction (RT-PCR) described by Takeshita et al. (2008).
To examine the adaptability of D3Yα on different host species, we used cucumber and tomato for inoculation assay (Table 1). For cucumber and tomato plants, when the fourth true leaf stage to expand, the second true leaf was inoculated with purified virus particles of CMV-Y (1 mg/ml) or with those of CMV-Y (1 mg/ml) plus in vitro transcripts of D3Yα. We inoculated three cucumber and 9 to 12 tomato plants in the first assay, and repeated the second assay by using different three cucumber and 7 to 8 tomato plants with the same inocula as described above (Table 1). At 28 days post inoculation (dpi), the upper, non-inoculated leaves were collected from cucumber and tomato plants and tested by RT-PCR as described by Takeshita et al. (2008).

Number of plants in which D3Yα was detected by RT-PCR using 3a-ORF-specific primers in upper, non-inoculated leaf tissues of total cucumber and tomato plants inoculated with CMV-Y virions (Y) or Y plus in vitro transcripts of D3Ya (Y + D3Ya)
To monitor the D3Yα generation through serial inoculation on N. benthamiana or N. tabacum, we mechanically inoculated the plants with in vitro transcripts of cDNA clones of CMV-Y RNAs1, 2, 3 as reported by Suzuki et al. (1991). As Kaplan et al. (2004) reported that N. tabacum cv. Samsun NN which was inoculated with in vitro transcripts of cDNA of CMV RNAs does not generate D RNA3, we used in vitro transcripts for the first inoculation. Those inoculated plants were designated as the first passage in Fig. 1A, and we collected upper, non-inoculated leaves at 10 dpi for sap inoculation in the second passage, and at 10 and 40 dpi for 3a-ORF-specific RT-PCR. For sap inoculation in the third passage, we further collected upper, non-inoculated leaves at 14 dpi in the second passage. For 3a-ORF-specific RT-PCR, we sampled upper, non-inoculated leaves at 14, 30, 90, and 180 dpi in the second and 14 and 30 dpi in the third passage (Fig. 1B and C).

Flow chart of steps for serial passages of strain Y of cucumber mosaic virus (CMV-Y) in Nicotiana tabacum, timing of sampling for polymerase chain reaction (PCR), and results of PCR to detect CMV-Y RNA3 and D3Yα in plants. (A) Flow chart shows the interval between inoculations for each passage, sampling times for 3a-ORF-specific reverse transcription PCR (RT-PCR), and PCR result for the presence (+) or absence (−) of D3Yα with RNA3. For each passage, two assay sets of three plants each were inoculated. In the first passage, plants were inoculated with in vitro transcripts derived from infective cDNA clones of CMV-Y RNAs 1, 2, and 3. At 10 days after inoculation (dpi), plants for the second passage were inoculated with sap from the first passage plants. At 14 dpi of second-passage plants, plants for the third passage were inoculated with sap from the second-passage plants. Plants were sampled for 3a-ORF-specific RT-PCR at the indicated times to determine when D3Yα was first detected and how long it was retained. (B, C) PCR results for D3Yα in samples at (B) 14, 30, 90, and 180 dpi in the second passage and at (C) 14 and 30 dpi in the third passage. M, DNA ladder marker; mc, mock control, plant was inoculated with phosphate buffer (pH 7.0); lanes 1-6, sample numbers for the two assay sets (1–3, 1st assay; 4–6, 2nd assay); however, sample (biological replicate number) 1 in the 1st assay of the second passage did not survive through 180 dpi. The presence of D3Yα is marked as + as in (A).
To investigate the involvement of temperature in generation or retention of D3Yα, N. benthamiana plants were inoculated with D3Yα-free purified CMV-Y virions or CMV-Y virions containing D3Yα, then those plants were kept at 15°C or 36°C till sampling at 7 and 28 dpi. The inoculum was prepared using tobacco plants which were inoculated with D3Yα-free CMV-Y virions (in the first passage as described above) at 7 to 10 dpi for checking the generation of D3Yα (Fig. 2A). Also, we prepared sap from N. tabacum plants that were inoculated with CMV-Y virions (1 mg/ml) plus in vitro transcripts of D3Yα for the retention of D3Yα at 15°C or 36°C (designated a positive control in Fig. 2A). We inoculated the second passage plants with sap from upper, non-inoculated leaves of the infected N. tabacum plants in the first passage. For 3a-ORF-specific RT-PCR analysis, upper, non-inoculated leaves of the second passage kept at 15°C were sampled at 28 dpi while we collected upper, non-inoculated leaves of the plants kept at 36°C at 7 dpi because the plants could not survive beyond 10 dpi.
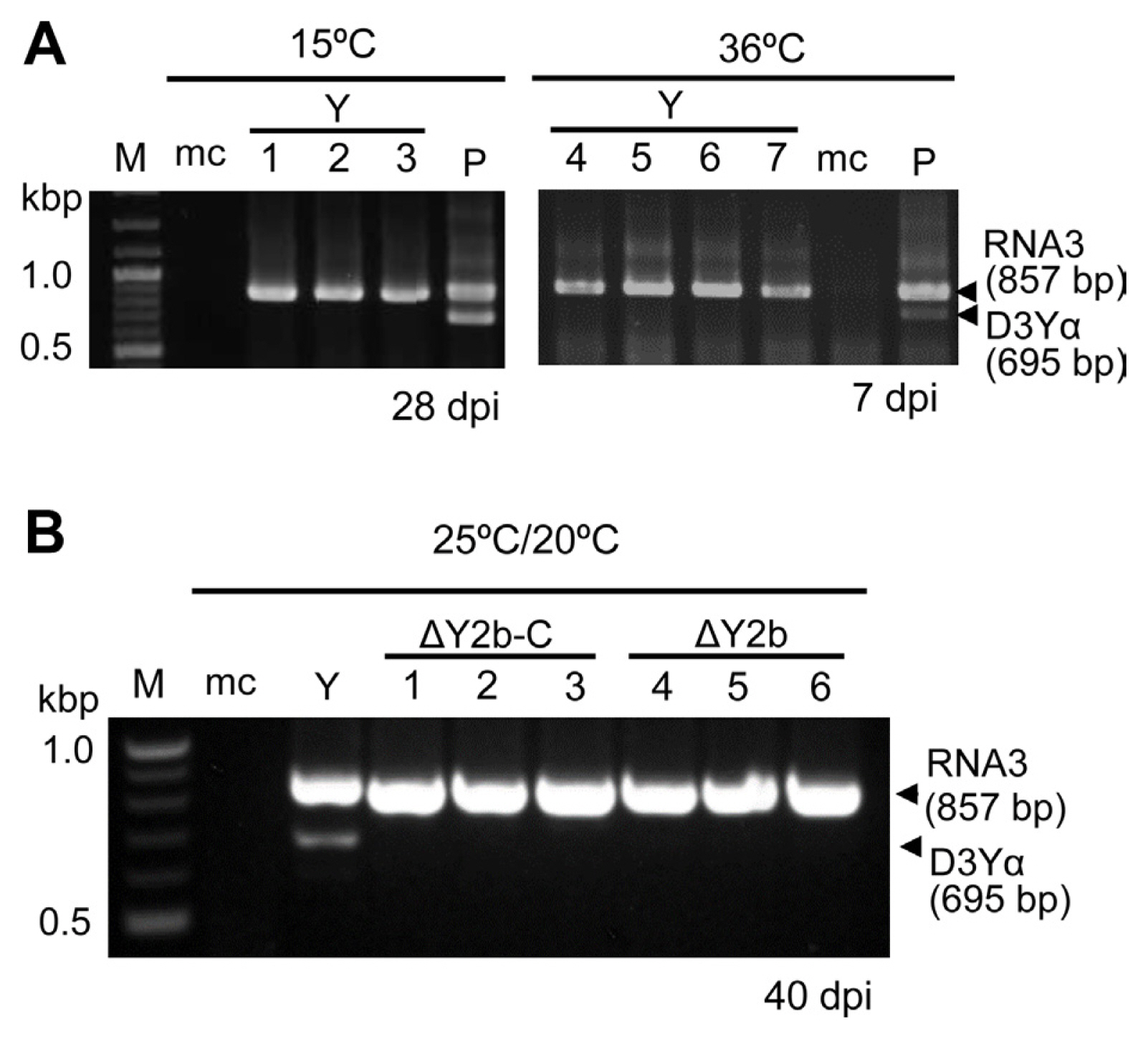
Results of 3a-OFR-specific reverse transcription polymerase chain reaction (RT-PCR) to detect D3Yα and RNA3 in upper, non-inoculated leaves of Nicotiana benthamiana grown at different temperatures and on different days after inoculation (dpi). (A) Plants were inoculated with purified strain Y of cucumber mosaic virus (CMV-Y, Y) virions (1 mg/ml) with/without D3Yα, then grown at 15°C or 36°C. M, DNA ladder marker; mc, mock control, plant inoculated with phosphate buffer (pH 7.0); P, a plant inoculated with CMV-Y virions plus D3Yα as positive control; lanes 1–3, sample plants grown at 15°C and sampled at 28 dpi; lanes 4–7, plants grown at 36°C and sampled at 7 dpi. (B) Plants were grown at 25°C/20°C (day/night) and sampled at 40 days after inoculation with a CMV-Y mutant. Y, a CMV-Y-infected N. benthamiana in the second passage; lanes 1–3, plants inoculated with ΔY2b-C mutant (C-terminus of 2b protein was truncated); lanes 4–6, plants inoculated with ΔY2b mutant (entire region of 2b protein with partially overlapped C-terminus of the 2a protein was deleted). The entire experiment was done twice (data not shown).
To examine whether the 2b protein is necessary for accumulating D3Yα, N. benthamiana plants grown at 25°C/20°C (day/night) were inoculated with CMV-Y or two CMV-Y deletion mutants (C2-A1 and C2-H1) as described by Takeshita et al. (2012). C2-A1 vector (ΔY2b-C in Fig. 2B) truncated C terminus of the 2b protein, and C2-H1 (ΔY2b in Fig. 2B) has the entire region of the 2b protein deleted including ORF region which partially overlaps with C terminus of the 2a protein (Supplementary Fig. 1). For the first passage, each in vitro transcript using cDNA clone of CMV-Y RNA1,3 and C2-A1/C2-H1 vector were prepared as described by Takeshita et al. (2012). Then, upper, non-inoculated leaf tissues in the first passage were collected at 10 dpi for sap inoculation in the second passage. We sampled upper, non-inoculated leaves in the second passage at 40 dpi for 3a-ORF-specific RT-PCR analysis.
RNA extraction and RT-PCR
Total RNA was extracted from leaves using RNAiso Plus (TaKaRa Bio, Kusatsu, Japan) according to the manufacturer’s protocol. For cDNA synthesis, total RNA was diluted to 50–300 ng/μl and stored at −80°C until use. cDNA was synthesized using ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO, Osaka, Japan) according to the manufacturer’s protocol. Two microliters of the diluted total RNA was added to 4.0 μl of deionized distilled water, then incubated at 65°C for 5 min. After 2.0 μl of 4× DN Master Mix with gDNA Remover was added, tubes were incubated at 37°C for 5 min. Finally, 2.0 μl of 5× RT Master Mix II was added, and samples were incubated at 37°C for 15 min for reverse transcription, then at 95°C for 5 min to inactivate the reverse transcriptase. The synthesized cDNA was stored at −30°C until use. For the PCR, 2 μl of cDNA mixture was used in a PCR mixture containing 5.0 μl of 2× PCR buffer, 2.0 μl of 2 mM dNTPs, 0.3 μl of 10 μM forward and reverse primers, 0.2 μl of deionized distilled water, and 0.2 μl of KOD Fx Neo (TOYOBO) polymerase. The 3a-ORF-specific RT-PCR was used to detect D3Yα as described by Takeshita et al. (2008) and yield amplicons of 857 bp for RNA3 and 695 bp for D3Yα. The thermalcycling conditions were 94°C for 2 min; 30 cycles of 94°C for 30 s, 58°C for 30 s, and 68°C for 2.5 min; 72°C for 2 min. The PCR products were resolved using 1.0% agarose gel electrophoresis.
Nucleotide sequencing
The nucleotide sequences of 3a-ORF-specific RT-PCR products were determined using the Big Dye Terminator DNA Sequencing Kit v. 3.1 (Applied Biosystems, Waltham, MA, USA) and the ABI Prism 310 Genetic Analyzer. The sequence was analyzed using the program GENETYX-Win v. 15 (Genetyx Corp., Tokyo, Japan).
Reverse transcription quantitative polymerase chain reaction analysis
To quantify CMV-Y, ΔY2b-C, and ΔY2b, the CP-gene-targeting one-step quantitative RT-PCR (RT-qPCR) analysis was performed as previously reported by Ota et al. (2021). Two primer sets were used to amplify the CP gene and SAMD as a reference gene in N. benthamiana (Khaing et al., 2020; Liu et al., 2012). Obtained data was analyzed by ΔΔCt method and statistically processed by t-test (P < 0.05 or P < 0.01) in Microsoft Excel software.
Biological assay of active replication of CMV-Y
Cowpea develops local lesions against CMV-Y infection according to Kim and Palukaitis (1997). We grew cowpea at 25°C/20°C during the experiment period. Non-inoculated, upper leaves of three to four N. benthamiana that were inoculated with CMV-Y, ΔY2b-C, or ΔY2b during the second passage were collected at 7 and 28 dpi, then we used these tissues for sap inoculation onto the primary leaves of cowpea to observe the development of local lesions at 7 dpi. Also, we examined the active replication of CMV-Y from N. benthamiana held at 15°C at 7 or 28 dpi or held at 36°C at 7 dpi. The number of local lesions per leaf was counted and the average number of local lesions and standard errors were calculated, then were statistically processed by t-test (P < 0.05 or P < 0.01).
Results
Retention of D3Yα on different host species
To examine host adaptation of D3Yα in other plant species, we inoculated cucumber and tomato plants with purified CMV-Y virions or with a mixture of CMV-Y virions and D3Yα as described earlier (Table 1). RT-PCR using 3a-ORF-specific primers showed that when mixture of CMV-Y and D3Yα was used as the inoculum, the upper, non-inoculated leaves of cucumber plants had accumulated D3Yα by 28 dpi, but only two out of twenty tested tomato plants retained D3Yα throughout experiment 1 and 2 (Table 1). These results suggested that D3Yα varies in its adaptability to different plant host species. Interestingly, D3Yα was not detected in cucumber and tomato plants at 28 days after inoculation with only CMV-Y, suggesting that the generation of D3Yα depends on the host (Table 1).
Serial inoculation and D3Yα detection on Nicotiana spp
In our previous study, no subviral RNAs other than D3Yα were detected in total RNA extracted from CMV-Y-infected N. benthamiana and N. tabacum plants, which were inoculated with in vitro transcripts of CMV-Y RNAs mixed with D3Yα, for at least 5 passages at 7-to-10-day intervals (Takeshita et al., 2008). Using a mixture of CMV-Y RNA transcripts as inoculum to establish that D3Yα has been generated is a key step to elucidate the mechanisms involved in D3Yα generation during virus infection. To test for the generation of D3Yα experimentally, we first examined N. tabacum that were inoculated with CMV-Y RNA1 to RNA3 transcripts in the first passage and did not detect D3Yα using 3a-ORF-specific RT-PCR throughout the experiment (10 and 40 dpi) (data not shown).
We then serially inoculated tobacco plants (the second passage) with sap from N. tabacum in the first passage to examine generation and maintenance of D3Yα in the plants over a longer period after inoculation (Fig. 1A). In the second passage, although D3Yα was not detected at 14 dpi in N. tabacum plants, it was detected at 30 dpi (Fig. 1B). We detected D3Yα at 90 and 180 dpi in the second passage; however, more plants had no D3Yα over time (Fig. 1B). These results indicated that by several weeks after inoculation, D3Yα levels start to decline for the next several months (Fig. 1B). Like during the second passage at 14 and 30 dpi, the same phenomenon occurred in the third passage (Fig. 1C). D3Yα was similarly generated in N. benthamiana (data not shown). All of the D3Yα RT-PCR products from the second and the third passage had the same nucleotide sequence that was reported by Takeshita et al. (2008) (Supplementary Figs. 2 and 3).
Effect of temperature on generation of D3Yα
To test the influence of temperature other than 25°C/20°C (day/night) on the generation and retention of D3Yα in plants, we first tested the effect of 15°C using N. benthamiana, which were inoculated with sap from the first passage plants infected with CMV-Y or CMV-Y plus D3Yα, but only incubating the plants at 15°C during the second passage (Fig. 2A). CMV-Y-infected N. benthamiana kept at 15°C showed stunting but not yellow mosaic symptom (Supplementary Fig. 4). None of the plants inoculated with CMV-Y had D3Yα at 7 and 28 dpi (Fig. 2A). On the hand, the plants, which were inoculated with CMV-Y plus D3Yα and held at 15°C, had retained D3Yα by 28 dpi (Fig. 2A). In subsequent test of the effects of high temperature on generation and maintenance of D3Yα, N. benthamiana plants were inoculated with sap from the first passage plants infected with CMV-Y or CMV-Y containing D3Yα and kept at 36°C during the second passage. By 7 dpi, none of the CMV-Y-inoculated plants generated D3Yα (Fig. 2A), and none survived beyond 10 dpi (Supplementary Fig. 4). However, the plants, which were inoculated with CMV-Y containing D3Yα, retained D3Yα at 36°C for at least 7 dpi (Fig. 2A). Once D3Yα was present in the virus populations, then D3Yα seemed to be retained at a wide range of temperatures. The temperature conditions that are suitable for retaining D3Yα do not always seem to correspond with the temperature at which D3Yα is generated by CMV-Y in N. benthamiana.
Relevant of the 2b protein to D3Yα generation
The 2b protein of CMV counteracts the host RNA silencing and the SA-mediated antiviral resistances (Nakahara and Masuta, 2014), drives selection of interviral recombinants between CMV RNA1/2 and tomato aspermy virus RNA3 (Shi et al., 2008), and promotes unloading of CMV from vasculature into nonvascular cells (Takeshita et al., 2012). To assess the potential effects of the 2b protein on D3Yα generation, we inoculated N. benthamiana with the vector CMV2-A1 (Otagaki et al., 2006) or CMV-H1 (Matsuo et al., 2007). The CMV2-A1 vector (ΔY2b-C) has a functional 2b protein but the 3′ portion of its ORF was deleted; the CMV-H1 vector (ΔY2b) is a deletion mutant of CMV-Y that lacks the entire gene for the 2b protein (Supplementary Fig. 1). For achieving long-lasting systemic infection after inoculation with ΔY2b, we selected N. benthamiana as a host to compare infection after inoculation with CMV-Y, ΔY2b-C, and ΔY2b (Fig. 2B). The CMV-Y- and ΔY2b-C-infected plants showed severe yellow mosaic and green patches in upper, non-inoculated leaves at 7 and 28 dpi, respectively, while ΔY2b mutant induced a slight vein clearing throughout the experiment (Supplementary Fig. 5). The 3a-ORF-specific RT-PCR amplified DNA fragments from the entire region of the 3a ORF but did not produce any smaller ones from the ΔY2b-C or ΔY2b-infected plants even at 40 dpi during the second passage (Fig. 2B). To assess replication and accumulation levels of CMV-Y, ΔY2b-C, and ΔY2b, we mechanically inoculated cowpea leaves using sap from N. benthamiana held at 25°C/20°C and used RT-qPCR targeting CP gene of CMV-Y to measure relative viral levels in the upper, non-inoculated leaves from the infected N. benthamiana at 7 and 28 dpi (Fig. 3). The number of local lesions on primary leaves of cowpea plants, which were inoculated with sap from CMV-Y- or ΔY2b-C-infected N. benthamiana at 7 or 28 dpi, did not differ significantly (Fig. 3A). In addition, RT-qPCR targeting the CP gene of CMV-Y showed a significant difference in levels between CMV-Y and ΔY2b-C and between CMV-Y and ΔY2b at 7 dpi but not at 28 dpi (Fig. 3B). The number of local lesions on cowpea was positively correlated with viral levels in N. benthamiana (Fig. 3A and B). Also, we inoculated the primary leaves of cowpea with CMV-Y-infected N. benthamiana kept at 15°C or 36°C to exam CMV replication activity (Fig. 3C). The number of local lesions on the primary leaves of cowpea inoculated with sap from N. benthamiana held at 15°C indicated that CMV-Y was replicating without generating D3Yα at 7 and 28 dpi (Fig. 3C). Interestingly, at 7 and 28 dpi with CMV-Y, sap from N. benthamiana held at 15°C after inoculation induced more local lesions than those that were kept at 25°C/20°C (day/night) or 36°C (Fig. 3A and C). The results suggested that the intact 2b protein rather than a high level of virus, facilitates the generation of D3Yα.

Mean number of local lesions on cowpea leaves in assay for replication of cucumber mosaic virus (CMV) and mean relative levels of viral RNA in Nicotiana benthamiana. (A) Mean number of local lesions on cowpea leaves at 7 days after mechanical inoculation with sap from N. benthamiana at 7 and 28 days post inoculation (dpi) with strain Y of CMV (CMV-Y) or mutants ΔY2b-C or ΔY2b and corresponding (B) mean relative accumulation levels of CMV in the sap inoculum determined by reverse transcription quantitative polymerase chain reaction (RT-qPCR) that target CMV-Y coat protein (CP) gene (B). Three to four biological replicates of upper, non-inoculated leaves were collected from N. benthamiana plants, grown at 25°C/20°C, at 7 and 28 dpi for the sap inoculum of the primary leaves of cowpea and for RT-qPCR targeting CP gene of CMV-Y. (C) Mean number of local lesions on cowpea at 7 dpi, inoculated as in (A). Sap inoculum was prepared from N. benthamiana plants kept at 15°C or 36°C. For sap inoculation, leaves were homogenized in 2.5 times volume (v/w) of phosphate buffer (pH 7.0). (B) Relative levels of CMV-Y CP gene were standardized to SAMD transcript levels. Standard error bars are shown for each mean; means were analyzed for significant differences using a t-test (*P < 0.05; **P < 0.01; ns, not significant). nd: no data because the plants kept at 36°C did not survive after 10 dpi.
Discussion
In this study, CMV-Y generated defective RNA3 in N. tabacum after prolonged infection during the second and subsequent passages. Cloning and nucleotide sequencing of the generated subviral RNA confirmed that it was D3Yα (Supplementary Figs. 2 and 3). Serial passages with CMV-Y and prolonged virus infection are likely necessary for generating D3Yα in N. tabacum and N. benthamiana. The decreasing levels of D3Yα over time, however, suggested substantial inferiority or dispensability of D3Yα for long-term survival of CMV-Y RNAs in the same host plant. The kinetics of D3Yα in our experimental conditions suggest that potential bottlenecks hinder the generation and persistence of D3Yα in the fields. Szittya et al. (2003) reported that virus-induced gene silencing was suppressed at 15°C. Thus, suppression of the host RNA silencing at low temperatures is unlikely to facilitate production of D3Yα. Interestingly, by using the ΔY2b-C or ΔY2b vectors, we found a potential effect of the 2b protein rather than replication of CMV-Y in generating D3Yα. Perhaps the function of the C-terminus of the 2b protein is important for generation of D3Yα.
The experimental conditions that resulted in generation of D3Yα appeared to be generally similar to those needed for D RNA3s generation by other strains of CMV and brome mosaic virus (BMV) strains. The first CMV D RNA 3s, designated D RNA 3α and D RNA 3β, were documented by Graves and Roossinck (1995) and were produced after the fourth passage of CMV-Fny stock in N. tabacum and still retained after another passage. Takeshita et al. (2008) also reported the retention of D3Yα by the helper virus CMV-Y for at least five passages in N. tabacum and N. benthamiana. Kaplan et al. (2004) inoculated N. tabacum with a mixture of in vitro transcripts synthesized from cDNA clones of CMV-Fny RNAs and first detected D RNA 3-1 during the third passage. Interestingly, López et al. (2007) showed that D RNAs were generated from the Pl1 isolate of CMV in N. tabacum cv. Xanthi plants without any serial passages. Damayanti et al. (1999) found D-RNAs from BMV RNA3 in purified virus preparations and in barley plants at 8 weeks after inoculation with a mixture of BMV RNA transcripts. These D RNA3s have several common features in the length and the positions of the deleted sequences, and in the timing of appearance in their host plants. According to Ayllón et al. (1999), the similar features and kinetics of the D RNA3s in the different host plants suggested that common mechanisms (e.g., a template-switching event) are associated with the generation of D RNAs by plant viruses.
D3Yα retains the essential sequence signals for RNA synthesis and packaging, however, whether D3Yα generation is biologically significant after prolonged infection and how the molecular mechanisms function in the generation process remains uncertain. Takeshita et al. (2009) suggested that the presence of D3Yα resulted in down-regulation of RNA3 replication, because D3Yα accumulates to the same levels as the helper virus RNA3. Additionally, a D3Yα mutant carrying a green mosaic-inducible CP gene induced different symptoms from yellow mosaic induced by the wild-type CMV-Y in upper, non-inoculated leaves of N. tabacum, suggesting spatial competition between RNA3 and D3Yα in N. tabacum and N. benthamiana is a more likely mechanism during the replication of RNA3/D3Yα than suppression of total accumulation levels of viral and subviral RNAs (Takeshita et al., 2009). From a different point of view, subgenomic RNA4 from D3Yα could alternatively supply the CP for helper virus even though the packaging efficiency of D3Yα is much lower than that of helper virus RNA (Takeshita et al., 2009). Further analysis focused on the putative function of the 2b protein associated with the generation of D3Yα should lead to new insights into the various factors that influence viral RNA replication and selective packaging.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
This work was partially supported by JSPS KAKENHI grant number 14760029. This work was also supported by JST SPRING, Grant Number JPMJSP2105.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).