Differential Resistance of Radish Cultivars against Bacterial Soft Rot Caused by Pectobacterium carotovorum subsp. carotovorum
Article information
Abstract
Bacterial soft rot caused by Pectobacterium carotovorum subsp. carotovorum (Pcc) is one of the most severe diseases in radish cultivation. To control this plant disease, the most effective method has been known to cultivate resistant cultivars. Previously, we developed an efficient bioassay method for investigating resistance levels with 21 resistant and moderately resistant cultivars of radish against a strain Pcc KACC 10421. In this study, our research expanded to investigate the resistance of radish cultivars against six Pcc strains, KACC 10225, KACC 10421, ATCC 12312, ATCC 15713, LY34, and ECC 301365. To this end, the virulence of the six Pcc strains was determined based on the development of bacterial soft rot in seedlings of four susceptible radish cultivars. The results showed that the Pcc strains exhibited different virulence in the susceptible cultivars. To explore the race differentiation of Pcc strains corresponding to the resistance in radish cultivars, we investigated the occurrence of bacterial soft rot caused by the six Pcc strains on the 21 resistant and moderate resistant cultivars. Our results showed that the average values of the area under the disease progress curve were positively correlated with the virulence of the strains and the number of resistant cultivars decreased as the virulence of Pcc strains increased. Taken together, our results suggest that the resistance to Pcc of the radish cultivars commercialized in Korea is more likely affected by the virulence of Pcc strains rather than by race differentiation of Pcc.
Bacterial soft rot is a plant disease that causes significant economic losses because it occurs not only during the cultivation period but also during storage (De Boer and Kelman, 1978; Perombelon and Salmond, 1995). Bacterial soft rot in Brassica crops is mainly caused by Pectobacterium carotovorum subsp. carotovorum (Pcc, formerly Erwinia carotovora subsp. carotovora) and Pseudomonas marginalis pv. marginalis (Pmm); it has also been reported that several Pectobacterium spp. cause bacterial soft rot (Rimmer, 2007). In particular, as radish (Raphanus sativus) can be grown all year round and the area of continuous radish cultivation is increasing in Korea, the occurrence of bacterial soft rot in radish is gradually increasing (Korean Society of Plant Pathology, 2022). As a causal agent for bacterial soft rot in radish, the pathogenic bacteria Pcc, Pmm, and Pectobacterium chrysanthemi have been reported in Korea. Among them, Pcc is the most widely known pathogen, which is a gram-negative, phytopathogenic bacterium with a broad host range, including carrot, potato, and Chinese cabbage (Hahm et al., 1998; Korean Society of Plant Pathology, 2022; Park et al., 1999; Rimmer, 2007). Recently, soft rot disease caused by Pectobacterium spp. was reported in Korea (Hong et al., 2023; Park et al., 2023, 2023b, 2023c, 2023d, 2023e).
For the controlling bacterial soft rot, chemical control with Bordeaux solution and several antibiotics (Chung et al., 2003; Wright et al., 2005) and biological control with antagonistic microorganisms and bacteriophages (Cui et al., 2019; Jee et al., 2012; Shrestha et al., 2009; Tsuda et al., 2016) have been frequently used. However, these control methods have become ineffective because climate change led to more occurrences of bacterial soft rot in the radish (Jee et al., 2020; Lee et al., 2018b; Rimmer, 2007). Therefore, the cultivation of resistant varieties has been recognized as one of the most effective and environmentally friendly control methods for bacterial disease (Rimmer, 2007; Taylor et al., 2002; Vijeth et al., 2018; Yerasu et al., 2019). For developing resistant cultivars, disease screening experiments are necessary to effectively select a resistant individual from many seeds obtained through crossbreeding with a resistant genetic resource. Moreover, considering that resistance types such as qualitative or quantitative resistance may require different bioassay methods for the determination of disease resistance, it is essential to investigate the race differentiation of the pathogenic strains for disease resistance in crops.
Race differentiation of plant pathogens occurs when the host plant exhibits qualitative resistance against the pathogens (Enya et al., 2009; Hibberd et al., 1987; Hirai, 2006; James and Williams, 1980; Kuginuki et al., 1999; Risser et al., 1976; Takken et al., 1998; Yoshikawa, 1993). For example, the race of Fusarium oxysporum f. sp. melonis causing Fusarium wilt is determined by the presence or absence of Fom-1 and Fom-2 resistance genes; race 0 can only infect melon lacking both resistant genes; race 1 and 2 can infect melon cultivar containing the Fom-1 and Fom-2, respectively; and race 1.2 infects melon plants containing both resistance genes (Risser et al., 1976). In addition, there have been many reports of qualitative resistance in interactions of plants and pathogens; chili pepper against Xanthomonas euvesicatoria (Hibberd et al., 1987), Chinese cabbage against Plasmodiophora brassicae (Hirai, 2006; James and Williams, 1980; Kuginuki et al., 1999; Yoshikawa, 1993), and tomato against Fulvia fulva (Enya et al., 2009; Takken et al., 1998). Nevertheless, given that resistant radish cultivars have been commercialized and cultivated in the fields, the race differentiation of Pcc strains related to the resistance has not been reported.
In this study, we aimed (1) to explore the virulence of six Pcc strains based on the development of bacterial soft rot in seedlings of four susceptible radish cultivars, (2) to investigate the resistance level of 21 radish cultivars against six Pcc strains, and (3) to determine whether the resistance of radish cultivars is affected by the virulence of Pcc strains or by race differentiation of Pcc.
Materials and Methods
Plant and growth condition
Four radish cultivars sensitive to Pcc, ‘Songbaek’, ‘Boseokaltari’, and ‘Housecheongok’ from FarmHannong (Seoul, Korea), and ‘Keongangsiregi’ from Koregon (Anseong, Korea) were purchased and used to investigate the virulence of six Pcc strains (Lee et al., 2018b). To test the resistance degree of 21 commercial radish cultivars against 6 Pcc strains, ‘Taechong’ and ‘Kangchoo’ from Syngenta Korea (Seoul, Korea); ‘Kwandongyeorum’, ‘Bitgoeunyeolmoo’, ‘Janghyeongbom’, ‘Cheongunplus’, ‘Tamseureon’, and ‘YR Champion’ from Monsanto Korea (Seoul, Korea); ‘Kiljo’, ‘Superkiljo’, and ‘Mansahyoungtong’ from Nongwoobio (Suwon, Korea); ‘Baekchun’, ‘Chengdoo’, ‘Jangseng’, ‘Kaeuljeojang’, and ‘Dongha’ from Asia Seed (Seoul, Korea), ‘Awooriwoldong ’, ‘Jeonmuhoomu’, and ‘Kuekdong’ from Koregon (Anseong, Korea); and ‘Myeongkagaeul1ho’ and ‘Supersiregi’ from Jeil Seed (Jeungpyeong, Korea) were also purchased and used in the study. As a susceptible cultivar to Pcc, we used cultivars ‘Housecheongok’, ‘Boseokaltari’, and ‘Songbaek’. To measure the development of bacterial soft rot in radish seedlings, horticultural soil No. 2 (Punong, Gyeongju, Korea) was placed into a plastic pot (70 ml/pot; Bumnong, Jeongup, Korea). And then seeds of the radish cultivar were sown in the soil and cultivated in a greenhouse at 25 ± 5°C for 20 days.
Pectobacterium carotovorum subsp. carotovorum strains
The six Pcc strains (KACC 10225, KACC 10421, ATCC 12312, ATCC 15713, LY34, and ECC 301365) were used in this study. KACC 10225 and KACC 10421 were provided from the Korean Agricultural Culture Collection (KACC, Rural Development Administration, Jeonju, Korea), and ATCC 12312 and ATCC 15713 strains were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). LY34 and ECC 301365 strains isolated from kimchi cabbage and lettuce, respectively, were kindly provided by Dr. Seon Woo Lee, Dong-A University (Park et al., 1998). All Pcc strains were stored in a 20% glycerol solution at −75°C.
Virulence assay
To prepare inoculum, Pcc strains grown on nutrient agar (Becton, Dickinson and Company, Sparks, MD, USA) medium at 30°C for 24 h were inoculated into a 5-ml nutrient broth (NB; Becton, Dickinson and Company) for seed culture. The resulting culture was added into a new NB medium at a concentration of 1% and then incubated at 30°C with shaking (200 rpm). After 36 h incubation, the bacterial culture was centrifuged at 8,000 rpm and 4°C for 10 min (Beckman Coulter Inc., Brea, CA, USA), and the pellet of bacterial cells was resuspended in sterile water. The bacterial suspension was adjusted to an optical density of 0.2 at 600 nm (OD600), corresponding to 1.0 × 108 cells/ml, and then diluted (10−2).
A 5 ml of Pcc suspension (about 1.0 × 106 cells/ml) was drenched at the base of radish seedlings (3–4 fully expanded leaf-stage). The inoculated plants were incubated in a humid chamber at 25°C for 24 h, and then the plants were transferred and grown in a growth room (25°C and 80% relative humidity) with 12 h light per day (photosynthetic photon flux density: 55 μmol/m2·s).
Disease rating and statistical analysis
To assess the occurrence of bacterial soft rot in radish seedlings, the disease severity of each plant was investigated 2, 3, 4, and 5 days after inoculation of Pcc. Disease index (DI) was scored on a 0-to-4 scale, where 0 = no symptom; 1 = yellowing or soft rot symptom occurred in 1–25% of plant; 2 = yellowing or soft rot symptom occurred in 26–50% of plant; 3 = yellowing or soft rot symptom occurred in 51–75% of plant; and 4 = yellowing or softness symptoms occurred in 76–100% of plant, plant dead (Lee et al., 2018b). The response of radish cultivars against Pcc strains was interpreted by the DI values as resistance (R), ≤ 1.0; moderate resistance (MR), 1.1–2.0; and susceptibility (S), > 2.0 (Lee et al., 2018b). Disease severity (%) was calculated according to the following formula. Disease severity (%) = {∑(disease index × the number of diseased plants)/(4 × the number of plants rated)} × 100. The area under the disease progress curve (AUDPC) was also calculated according to the following equation (Jeger and Viljanen-Rollinson, 2001; Madden et al., 2007). AUDPC = ∑ n i = 1 [ t (i + 1) − ti] × [DS (i + 1) + DSi]/2; n = number of assessments, ti = number of days elapsed from the inoculation day to the assessment date i, DSi = disease severity (%) on assessment date i. All experiments were performed in 10 replicates with two runs. All data were subjected to a one-way analysis of variance (ANOVA) and the significance of treatments was determined by Duncan’s multiple range test (P = 0.05).
Results
Virulence of Pectobacterium carotovorum subsp. carotovorum strains
When four susceptible radish cultivars (‘Keongangsiregi’, ‘Boseokaltari’, ‘Songbaek’, and ‘Housecheongok’) were inoculated with six Pcc strains (KACC 10225, KACC 10421, ATCC 12312, ATCC 15713, ECC 301365, and LY34), the tested cultivars showed different responses in the extent of bacterial soft rot development depending on the Pcc strains, starting 3 days post-inoculation (dpi) (Fig. 1). At 4 dpi, radish seedlings of all the cultivars inoculated with ATCC 12312 and ECC 301365 were severely yellowed or dead; the disease severity values by ATCC 12312 were 95% for ‘Keongangsiregi’, 100% for ‘Boseokaltari’, 88% for ‘Songbaek’, and 100% for ‘Housecheongok’, with an average disease severity of 96%, and also the disease severity values by ECC 301365 were 93% for ‘Keongangsiregi’, 93% for ‘Boseokaltari’, 75% for ‘Songbaek’, and 100% for ‘Housecheongok’, with an average disease severity of 90% (Fig. 1). However, when inoculated with LY34, the disease severity values were 30% for ‘Keongangsiregi’, 25% for ‘Boseokaltari’, and 28% for both ‘Songbaek’ and ‘Housecheongok’, with an average disease severity of 28% (Fig. 1).
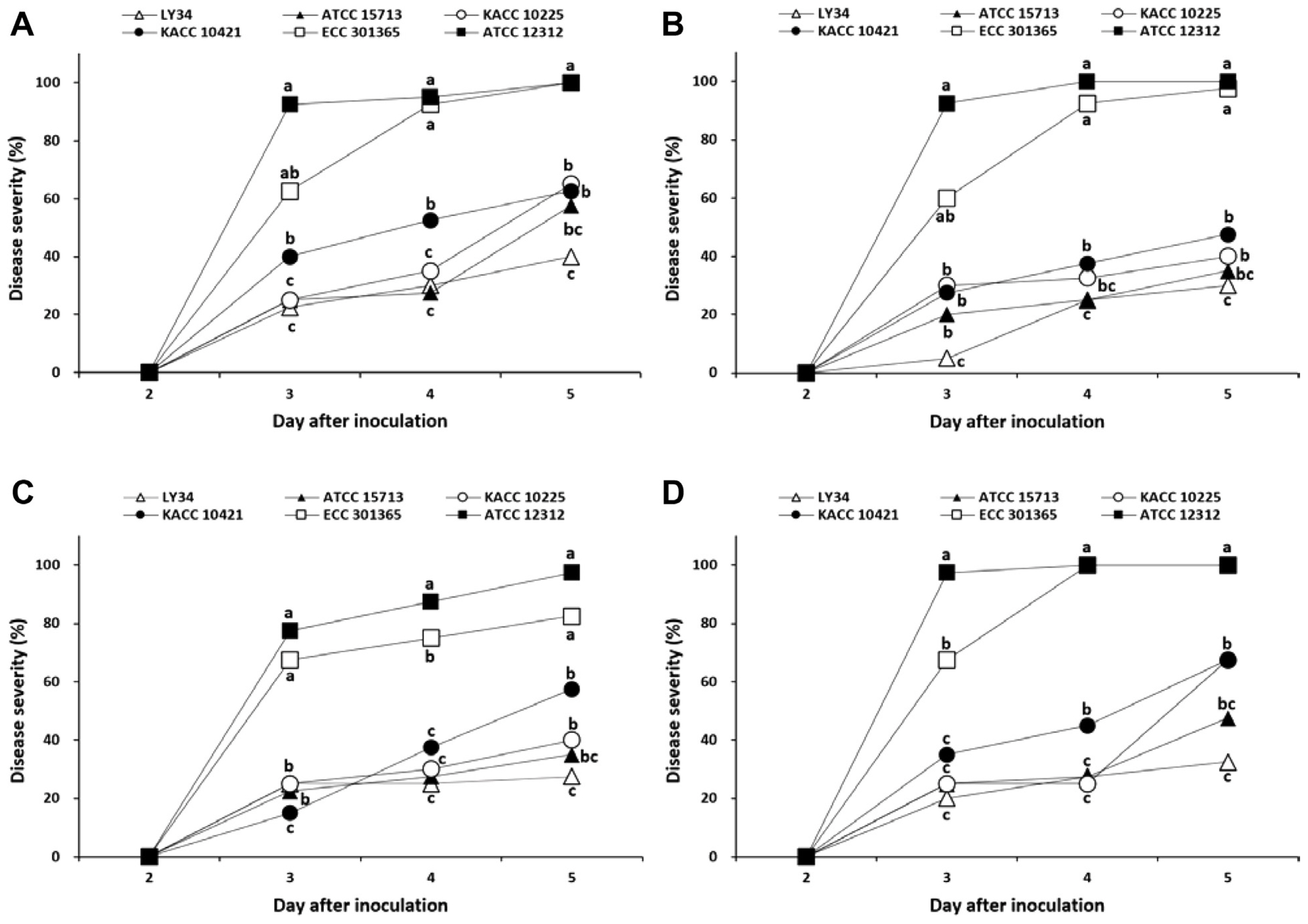
Disease severity of bacterial soft rot caused by Pectobacterium carotovorum subsp. carotovorum strains on susceptible radish cultivars. (A) ‘Keongangsiregi’. (B) ‘Boseokaltari’. (C) ‘Songbaek’. (D) ‘Housecheongok’. Twenty-day-old seedlings were inoculated by drenching the proximal part of plants with 5 ml bacterial suspension at a concentration of 8.0 × 105 cells/ml. The inoculated plants were incubated in a dew chamber at 25°C for 24 h, then transferred to a growth room at 25°C, 80% relative humidity, and 12 h of light exposure daily. At 2–5 days after inoculation, the disease severity of plants was investigated on a scale of 0–4, which was then converted to a percentage. Values labeled with the same letter within each day after inoculation are not significantly different based on Duncan’s multiple range test at P = 0.05.
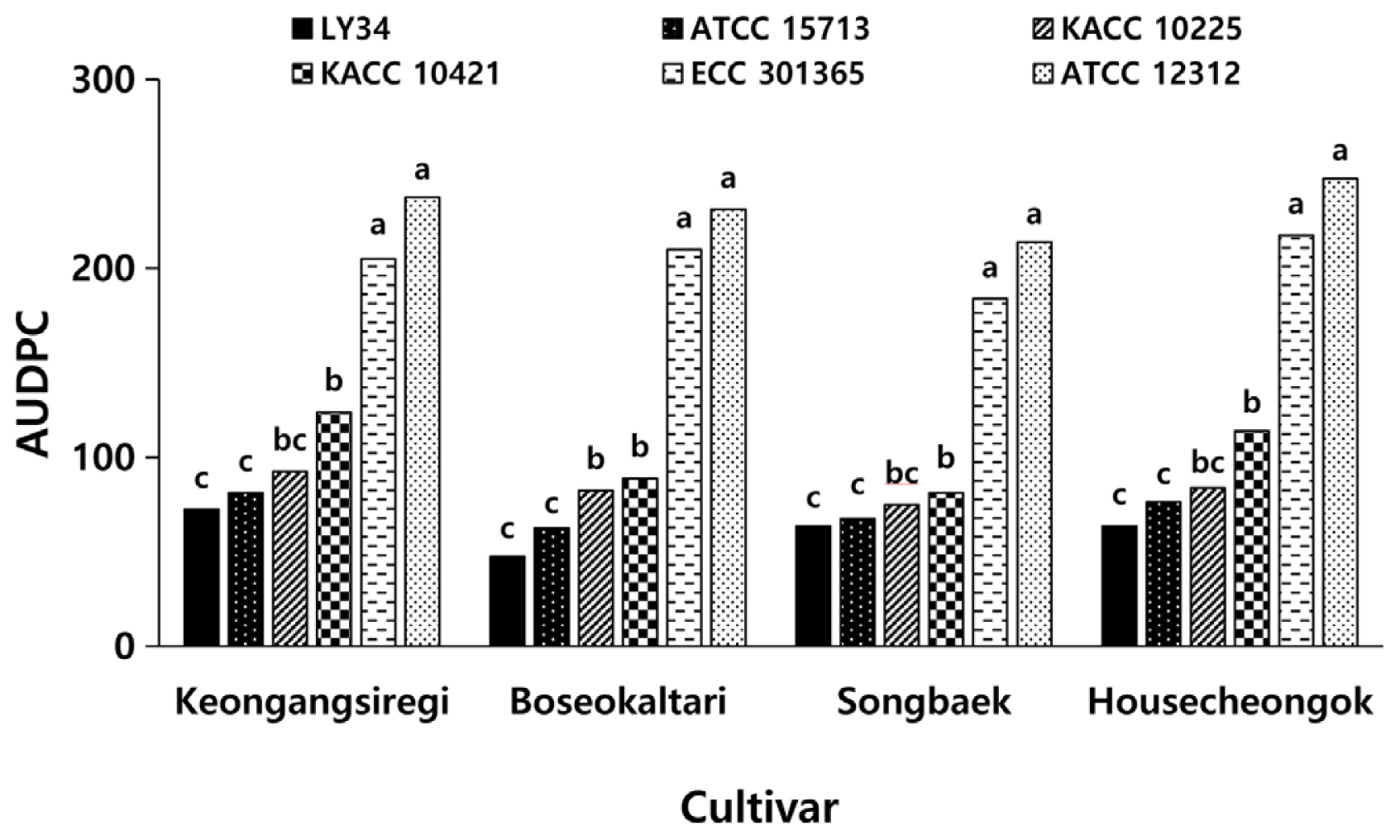
When we calculated the AUDPC based on the results of bacterial soft rot development in radish seedlings at 1, 3, 4, and 5 dpi, the average values of AUDPC for the six Pcc strains were 135 for ‘Keongangsiregi’, 120 for ‘Boseokaltari’, 114 for ‘Songbaek’, and 134 for ‘Housecheongok’ (Fig. 2). However, in terms of the pathogens’ virulence, radish cultivars inoculated with LY34 strain had the lowest AUDPC of 62, followed by ATCC 15713, KACC 10225, KACC 10421, ECC 301365, and ATCC 12312 with average AUDPC values of 72, 84, 102, 204, and 233, respectively (Fig. 2). These results suggest that among the Pcc strains used in the study, ATCC 12312 exhibited the strongest virulence, followed by ECC 301365, KACC 10421, KACC 10225, ATCC 15713, and LY34. Furthermore, statistical analysis of the AUDPC of bacterial soft rot on the four radish cultivars by each Pcc strain also showed that the Pcc strains can be divided into three groups based on their virulence; the most virulent group was ATCC 12312 and ECC 301365, the medium virulence group was KACC 10421 and KACC 10225, and the least virulent group was ATCC 15713 and LY34 strains (Fig. 2). Taken together, our results presented that there were no significant differences in the occurrence of bacterial soft rot among radish cultivars, but there were differences in virulence between Pcc strains.
Occurrence of bacterial soft rot in 24 radish cultivars caused by Pectobacterium carotovorum subsp. carotovorum strains
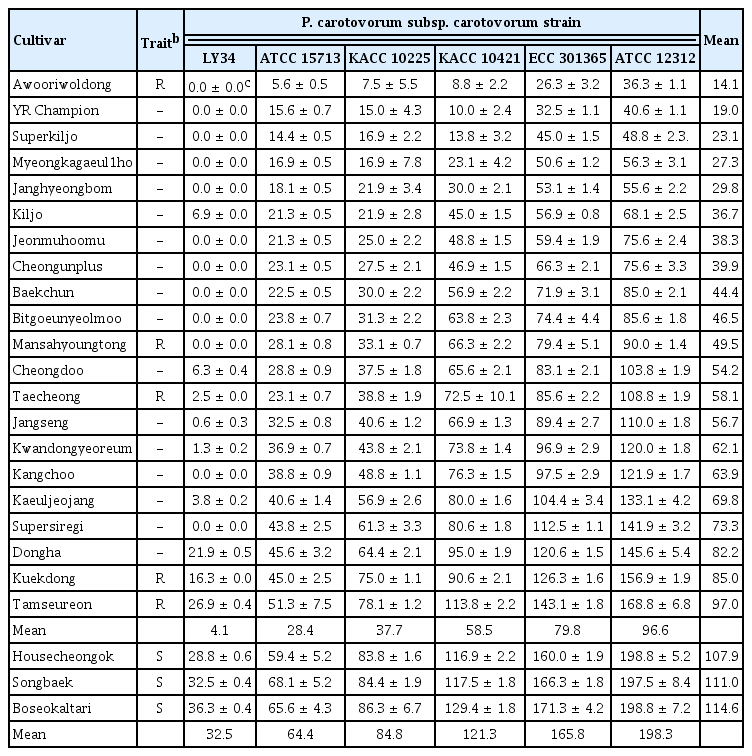
To investigate the occurrence of bacterial soft rot in different radish cultivars against six Pcc strains, we selected 24 radish cultivars: five radish cultivars (‘Awooriwoldong’, ‘Mansahyoungtong’, ‘Taechong’, ‘Kuekdong’, and ‘Tamseureon’) marketed as resistant cultivars to Pcc, 16 cultivars (‘YR Champion’, ‘Superkiljo’, ‘Myeongkagaeul1ho’, ‘Janghyeongbom’, ‘Kiljo’, ‘Jeonmuhoomu’, ‘Cheongunplus’, ‘Baekchun’, ‘Bitgeunyeolmoo’, ‘Cheongdoo’, ‘Kwandongyeoreum’, ‘Kangchoo’, ‘Kaeuljeojang’, ‘Supersiregi’, and ‘Dongha’) identified as resistant or moderate resistant cultivars to Pcc KACC 10421 based on our previous our study (Lee et al., 2018b), and three susceptible cultivars (‘Housecheongok’, ‘Songbaek’, and ‘Boseokaltari’). The average AUDPCs of bacterial soft rot in the resistant or moderate resistant cultivars against LY34, ATCC 15713, KACC 10225, KACC 10421, ECC 301365, and ATCC 12312 were 4.1, 28.4, 37.7, 58.5, 79.8, and 96.6, respectively (Table 1). Whereas, the average AUDPC in three susceptible varieties (‘Housecheongok’, ‘Songbaek’, and ‘Boseokaltari’) against LY34, ATCC 15713, KACC 10225, KACC 10421, ECC 301365, and ATCC 12312 were 32.5, 64.4, 84.8, 121.3, 165.8, and 198.3, respectively (Table 1). Among the 21 resistant and moderate resistant cultivars, ‘Awooriwoldong’ exhibited the lowest occurrence of bacterial soft rot with an average AUDPC value of 14.1 against all Pcc strains followed by ‘YR Champion’, ‘Superkiljo’, ‘Myeongkagaeul’, ‘Janghyeongbom’, ‘Kiljo’, ‘Jeonmuhoomu’, ‘Cheongunplus’, ‘Baekchun’, ‘Bitgoeunyeolmoo’, ‘Mansahyoungtong’, ‘Cheongdu’, ‘Taecheong’, ‘Jangseng’, ‘Kwandongyeoreum’, ‘Kangchoo’, ‘Kaeuljeojang’, ‘Supersiregi’, ‘Dongha’, ‘Kuekdong’, and ‘Tamseureon’ (Table 1). Together, our results presented that the incidence of bacterial soft rot in the radish cultivars increased in proportion to the virulence of the six Pcc strains without specific resistance responses between the resistant cultivars and Pcc strains.
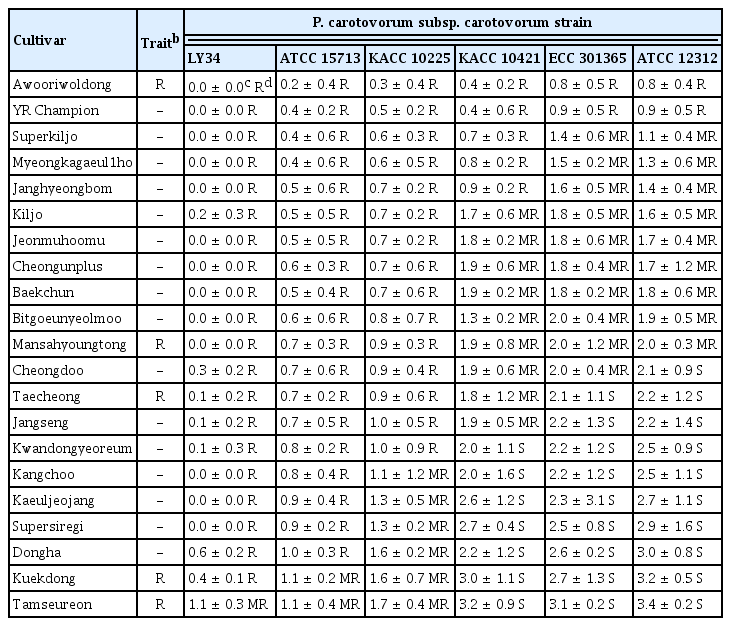
In terms of plant-resistant responses, the disease severity of the 21 resistant and moderate resistant radish cultivars against Pcc strains was investigated at 5 dpi. Two cultivars (‘Awooriwoldong’ and ‘YR Champion’) exhibited highly resistant responses against all Pcc strains, and nine cultivars (‘Superkiljo’, ‘Myeongkagauel1ho’, ‘Janghyeongbom’, ‘Kiljo’, ‘Jeonmuhoomu’, ‘Cheongunplus’, ‘Baekchun’, ‘Bitgoeunyeolmoo’, and ‘Mansahyoungtong’) were resistant or moderate resistant against all strains. The remaining 10 cultivars showed resistance or moderate resistance against at least one Pcc strain (Table 2). In other words, among the 21 radish cultivars tested, ‘Awooriwoldong’ and ‘YR Champion’ represented resistance response to all Pcc strains tested, but the resistance degree of the other cultivars against the Pcc strains was negatively correlated with the virulence of the bacterial strains (Table 2, Fig. 2). In the case of a cultivar ‘Chengdoo’, the cultivar was resistant against LY34, ATCC 15713 and KACC 10225, which are less virulent, moderate resistant to KACC 10421 and ECC 301365, and susceptible against ATCC 12312, which is the most virulent (Table 2, Fig. 2). A cultivar ‘Kangchoo’ was resistant against LY34 and ATCC 15713, which are the least virulent pathogens, moderate resistant against KACC 10225, and susceptible against KACC 10421, ECC 301365, and ATCC 12312, which is relatively virulent (Table 2, Fig. 2). When we counted the number of cultivars showing resistance against each of the six Pcc strains, among the 21 radish cultivars, the number of cultivars against LY34, ATCC 15713, KACC 10225, KACC 10421, ECC 301365, and ATCC 12312 were 20, 19, 15, 5, 2, and 2, respectively (Table 3). Whereas, the number of cultivars showing susceptible responses against strains LY34, ATCC 15713, KACC 10225, KACC 10421, ECC 301365, and ATCC 12312 were 0, 0, 0, 7, 9, and 10, respectively (Table 3). Thus, these results indicated that the resistance degree of the tested radish cultivars is negatively correlated with the virulence of the Pcc strains.
Discussion
It has been reported that the strains of various plant pathogens exhibited different virulence. For example, Jo et al. (2016) showed that of the four P. brassicae strains, the YC strain caused the most severe clubroot in the cabbage seedlings, followed by GN1, HN1, and DJ strains, when 16 susceptible cabbage cultivars were inoculated by the P. brassicae strains. Lee et al. (2018a) also investigated the occurrence of bacterial wilt on 12 chili pepper cultivars by inoculating with 14 isolates of Ralstonia solanacearum and reported that the R. solanacearum could be classified into six groups according to their virulence. In addition, among eight isolates of Fusarium oxysporum f. sp. raphani causing Fusarium wilt on radish, JHW isolate had the strongest virulence on radish seedlings, followed by HN, NW1, KR1, 147A, 57A, 59A, and 60A isolates, and these isolates could be divided into three groups based on their virulence on radish seedlings (Lee et al., 2020). In this study, we investigated the virulence of Pcc strains on four susceptible radish cultivars, and our results showed that of six Pcc strains, ATCC 12312 exhibited the strongest virulence followed by ECC 301365, KACC 10421, KACC 10225, ATCC 15713, and LY34, supporting that there were differences in virulence between the Pcc strains.
Additionally, we examined the disease response of 21 radish cultivars against 6 Pcc strains. The occurrence of bacterial soft rot in each cultivar presented a positive correlation with the virulence of the Pcc strains, and the average disease severity of all tested radish cultivars increased as the virulence of the Pcc strains increased (Tables 1 and 2, Fig. 2). Moreover, the radish cultivars exhibited resistance in inverse proportion to the virulence of the inoculated Pcc strain (Tables 2 and 3, Figs. 1 and 2). In particular, we did not observe a specific response between the resistant cultivars and Pcc strains, supporting the qualitative resistance (Hibberd et al., 1987; Kim et al., 2016; Kuginuki et al., 1999; Ramirez-Villupadua et al., 1985; Risser et al., 1976). Therefore, our results suggest that the resistance of the radish cultivars against Pcc strains used in this study seems to be quantitative, not qualitative.
During the developing process of resistant cultivars, it is essential to effectively select a resistant individual through disease screening experiments determined by crop/pathogen interactions such as qualitative or quantitative resistance. Given that crop/pathogen interaction results in qualitative resistance, race differentiation of pathogenic isolates to the resistance genes of the host plant occurs. In other words, to develop a resistance cultivar, a specific race of pathogens corresponding to a host resistance gene should be used in a bioassay. For example, the race of F. oxysporum f. sp. melonis causing Fusarium wilt is determined by the presence or absence of Fom-1 and Fom-2 resistance genes (Risser et al., 1976). Therefore, to develop resistant melon cultivars containing the Fom-1 and Fom-2, it is necessary to use a corresponding race to Fom-1 and Fom-2 for the breeding.
In contrast, in the case of quantitative resistance, there is no race differentiation of pathogenic fungi and bacteria to resistance genes of resistant plants, and the plants exhibit resistance in inverse proportion to the virulence of the pathogenic strains. For example, the resistant chili pepper cultivars did not exhibit a specific resistant response against Phytophthora capsici isolates, and the resistance degree of the chili pepper is affected by the virulence of the pathogen (Foster and Hausbeck, 2009; Jo et al., 2014; Yang et al., 1989). Lee et al. (2018a) also showed that the resistance of 12 chili pepper cultivars against 14 R. solanacearum strains was negatively correlated with the virulence of the bacterial pathogens, and 14 R. solanacearum strains can be divided into six groups based on their virulence. Thus, unlike the case of qualitative resistance, the screening experiment to breed plants for quantitative resistance requires the selection of strains with appropriate virulence according to the research purpose.
Here, we found that the occurrence of bacterial soft rot on the seedlings of radish cultivars is positively correlated with the virulence of the strains. Thus, the resistance of commercial radish cultivars in Korea is more likely affected by the virulence of Pcc strains rather than by race differentiation of Pcc.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through the Digital Breeding Transformation Technology Development Program funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) with a grant number: 322059-03-1-HD060.