 |
 |
| Plant Pathol J > Volume 32(3); 2016 > Article |
Abstract
A bacterial tyrosine sulfotransferase, RaxST, is required for activation of rice XA21-mediated immunity, and it catalyzes sulfation of tyrosine residues of Omp1X and RaxX in Xanthomonas oryzae pv. oryzae, a causal agent of bacterial blight in rice. Although RaxST is biochemically well-characterized, biological functions of tyrosine sulfation have not been fully elucidated. We compared protein expression patterns between the wildtype and a raxST knockout mutant using shotgun proteomic analysis. Forty nine proteins displayed a more than 1.5-fold difference in their expression between the wildtype and the mutant strains. Clusters of orthologous groups analysis revealed that proteins involved in cell motility were most abundant, and phenotypic observation also showed that the twitching motility of the mutant was dramatically changed. These results indicate that tyrosine sulfation by RaxST is essential for Xoo movement, and they provide new insights into the biological roles of RaxST in cellular processes.
Xanthomonas oryzae pv. oryzae (Xoo), a Gram-negative bacterium, motile by means of a single polar flagellum, produces polysaccharide xanthan gum, with its characteristic yellow pigments, and also causes bacterial blight disease in rice plants (Mew, 1987). Blight disease is one of the most destructive diseases plaguing rice in Africa and Asia, and the losses caused by this disease account for those of up to half of rice production worldwide, resulting in tremendous economic and social impact (Gnanamanickam et al., 1999). In addition to its importance, the bacterium is a model organism for studying the function of genes and proteins because its full genome has been sequenced and it has been studied with diverse well-established analytical techniques (Han et al., 2008; Park et al., 2014b; Salzberg et al., 2008).
Tyrosine sulfation is one of the most common posttranslational modifications in eukaryotic organisms, and up to 1% of tyrosine residues on total proteins can be sulfated in eukaryotic organisms (Monigatti et al., 2006). Tyrosine sulfation is mediated by tyrosine sulfotransferases, which transfer a sulfate group (SO3−) from the universal sulfate donor, 3′-phosphoadenosine-5′-phosphosulfate, to specific substrates, and it occurs as an enzymatic modification of peptides and proteins. In contrast to serine/threonine phosphorylation, which is often involved in signal transduction pathways, tyrosine sulfation modulates protein-protein/peptide or ligand-receptor interactions with high affinity (Stone et al., 2009). A notable example in plants is phytosulfokine-α (PSK), which possesses sulfate tyrosine residues and serves as a self-recognition factor for the proliferation of plant cells (Matsubayashi et al., 2002). PSK is recognized by the phytosulfokine receptor kinase present in the plant plasma membrane, and tyrosine sulfation of PSK is required for ligand-receptor recognition. In animals, sulfation of tyrosine residues on the CC-chemokine receptor 5 is critical for both the binding of glycoprotein 120 from human immunodeficiency virus (HIV) and for the entry of HIV into human cells (Farzan et al., 1999). However, tyrosine sulfotransferases in prokaryotic organisms have not been well described compared to their counterparts in eukaryotes.
RaxST protein from Xoo is required for the triggering of XA21-mediated immunity and it shows homology with sulfotransferases, including human tyrosylprotein sulfotransferase 1 (TPST1) and TPST2, as well as Sinorhizobium meliloti nodulation protein H (da Silva et al., 2004). In the central region of the protein, RaxST possesses the 5′-phosphosulfate-binding and 3′-phosphate-binding motifs that are hallmarks of sulfotransferases and critical for the binding of 3′-phosphoadenosine-5′-phosphosulfate. Recently, the tyrosine sulfotransferase activity of RaxST was demonstrated using a novel sulfotransferase activity assay and ultraviolet photodissociation mass spectrometry (Han et al., 2012). RaxST specifically catalyzes the sulfation of tyrosine residues on both Omp1X (outer membrane protein 1 in Xoo) and a synthetic N-terminal peptide of CC-chemokine receptor 5. Omp1X is involved in Xoo motility and biofilm formation (Park et al., 2014b). Comparative proteomic analysis with an Omp1X knockout strain (XooΔomp1X) showed that the expression of 106 proteins was significantly changed. Clusters of orthologous groups (COG) analysis revealed that proteins related to cell motility and signal transduction were most abundant. Phenotypic observation also displayed that pili- and flagella-dependent motility of XooΔomp1X was significantly impaired. In addition, RaxST transfers sulfuryl groups to RaxX protein that is an activator of XA21-mediated immunity (Pruitt et al., 2015).
Although RaxST is the first biochemically characterized prokaryotic tyrosine sulfotransferase, it is still unclear which biological processes are associated with RaxST in Xoo. In this study, we carried out a label-free comparative shotgun proteomic analysis and a COG analysis in order to understand the biological processes regulated by RaxST in Xoo. Our results indicate that RaxST is involved in Xoo motility. Phenotypic observation also confirmed that RaxST is indispensable for Xoo motility. These results allow us to elucidate protein functions via a combination of proteomic analysis and phenotypic changes in Xoo.
We used the wildtype Xoo strain Philippines race 6, PXO99Az, and the raxST knockout mutant (XooΔraxST) that have been employed in previous studies (da Silva et al., 2004; Hopkins et al., 1992). To generate a complemented strain XooΔraxST (RaxST), the raxST gene was amplified using the forward primer 5′-CTCGAGATGCACCACCACCACCACCACGCTTGGCTGGCGATGCGTCCCGCCG-3′ and the reverse primer 5′-CTTCGCATGGAGCGCTGCTGGTATGAAGCTT-3′. The amplified DNA fragment was cloned into pTOP blunt V2 vector (Invitrogen, Carlsbad, CA, USA) and the cloned DNA, cut out with XhoI and HindIII, was inserted into a broad host range vector, pBBR1-MCS5, digested with same restriction enzymes (Kovach et al., 1995). The construct was introduced into XooΔraxST by electroporation. The complimented strain XooΔraxST (RaxST) successfully triggered XA21-mediated immunity in the rice cultivar Kitaake expressing the XA21 protein, but not in XooΔraxST (Fig. S1). These results indicate that the interruption of the raxST gene does not have any polar activity, which is consistent with a previous report (da Silva et al., 2004).
To understand the biological processes associated with RaxST, we compared protein expression between the wildtype and XooΔraxST using a label-free shotgun proteomic approach following previously established protocols (Park et al., 2014a). Briefly, Xoo strains were grown in peptone sucrose broth to 0.6 OD at 600 nm, and harvested. Bacterial cells were dissolved in 1 ml lysis buffer (6 M guanidine-HCl, 50 mM Tris-HCl, and 10 mM DTT; pH 7.8), and then were disturbed by sonication using an Ultrasonic Processor (Cole Parmer, Niles, IL, USA). Extracted proteins (1 mg) were precipitated using trichloroacetic acid and then digested with trypsin (Promega, Madison, WI, USA). The trypsin-digested protein (2 μg) was examined with an LTQ Velos Pro dual-pressure linear ion trap mass spectrometer combined with a split-free nano liquid chromatograph (EASY-nLC II; Thermo Fisher Scientific, Waltham, MA, USA) in nanospray ionization mode. The samples were separated in a column composed of MAGIG C18AQ 200A (5 μm; Michrom Bioresources, Auburn, CA, USA) and the peptides were eluted. Full MS spectra were gained with six data-dependent MS/MS scans (m/z 300-2,000 mass range). Three biological replicates from each strain were carried out.
To interpret the MS/MS spectra, we have used previously reported methods (Park et al., 2014a, 2014b), combination of a Thermo Proteome Discoverer 1.3 (ver. 1.3.0.399; Thermo Fisher Scientific) and the SEQUEST search algorithm was employed. The spectra were analyzed using the Xoo PXO99Az reference sequence database containing 4,375 proteins, from National Center for Biotechnology Information. Trypsin was situated as a cleavage enzyme, and up to two missed cleavages were allowable. All peptides had a 0.01 false discovery rate with forwarded and reversed database searches. One hundred ppm of precursor mass accuracies were acceptable and scores of probability from all peptides were over 20. The methionine oxidation was reflected as a possible modification. Proteins that matched at least two unique peptides were appreciated to be present in all samples. For comparative analysis of protein expression between the wildtype and XooΔraxST, a peptide-spectrum match (PSM) was used (Choi et al., 2008). PSMs from individual proteins were normalized against the total number of PSMs from the total proteins in each sample. Proteins shared in three biological replicates were chosen and used for comparison. The coefficient of variation calculated with all conserved proteins in three biological replicates was used for quality assessment. The mean value of the normalized PSMs was calculated per each protein and employed as a comparison value between the wildtype and XooΔraxST to identify differently expressed proteins. Proteins whose expression has shown over 1.5-fold difference between the wildtype and XooΔraxST were selected as differently expressed proteins.
In the three biological replicates with the wildtype strain, 753, 771, and 779 proteins were detected from 57,274, 53,045, and 60,401 PSMs, respectively. In the XooΔraxST strain, 768, 782, and 781 proteins were identified from 56,642, 57,027, and 60,071 PSMs, respectively (Table 1). Among the identified proteins, 661 and 662 proteins commonly detected in all three biological replicates of the wildtype and XooΔraxST strains, respectively, were used for comparative analysis. Counts of PSMs from 661 and 662 proteins from the wildtype and XooΔraxST strains, respectively, were compared after normalization. The average of coefficient of variation from all shared proteins in the wildtype and XooΔraxST strains was 15.88 and 16.12, respectively, suggesting that the degree of variation from means of PSMs is relatively low and the comparative analysis shows high confidence. The expression of a total of 49 proteins was affected by the presence of functional RaxST (Fig. 1A). Among these differentially expressed proteins, 22 and 27 proteins were only detected or highly expressed (over 1.5 fold) in the wildtype and XooΔraxST strains, respectively. These results indicate that approximately 7% of the observed proteins were specifically influenced by tyrosine sulfation by RaxST in Xoo. This is a relatively low number compared with phosphorylation by kinases on response regulators of bacterial two-component systems, leading to massive changes in gene expression (Mitrophanov and Groisman, 2008). Relatively low effects on gene expression by tyrosine sulfation in Xoo are consistent with a result observed in eukaryotic cells in which tyrosine sulfation is mostly involved in protein-protein/peptide or receptor-ligand interaction with high affinity, rather than in gene expression for cell signaling cascades via phosphorylation by kinases (Kehoe and Bertozzi, 2000).
Next, we carried out COG analysis (Tatusov et al., 2000) to classify 49 RaxST-associated proteins according to their predicted functions (Table 2, 3). The majority of proteins whose expression is affected by RaxST belong to the “cell motility” (N) and “signal transduction mechanisms” (T) categories (Fig. 1B). Eight of ten proteins belonging to “cell motility” are also categorized in “signal transduction mechanisms” (Table 3, included in other group T). Interestingly, eight chemotaxis-related proteins associated with twitching motility were identified as proteins altered by RaxST, and all of them (PXO_00057, PXO_00050, PXO_00047, PXO_00046, PXO_00032, and PXO_06212 from “cell motility”; PXO_06210 and PXO_00031 from “signal transduction mechanisms”) were detected only in XooΔraxST but not in the wildtype strain under the given conditions (Table 2, 3). In agreement with these observations, a comparative proteomic analysis with an Omp1X-knockout mutant (XooΔomp1X) also displayed differential expression of chemotaxis-related proteins (Park et al., 2014b). These results suggest that tyrosine sulfation by RaxST is most likely associated with bacterial movement. In addition to bacterial motility, the expression of 11 proteins classified as “function unknown” (S) was also altered (Table 2, 3), suggesting that RaxST might play a role in uncharacterized biological processes.
Bacterial chemotaxis is closely related to twitching motility in terms of pili-dependent motility (Miller et al., 2008). Overexpression and knockout mutations of genes involved in chemotaxis mechanisms resulted in impaired pili-dependent twitching motility (Kearns et al., 2001; Whitchurch et al., 2004). Because a proteomic analysis with XooΔraxST also implies that RaxST seems to have an effect on bacterial movement, the twitching motility of the wild type, XooΔraxST, and the complemented strain XooΔraxST (RaxST) was examined on potato sucrose agar (PSA) media containing 1.5% agar (Fig. 2). A twitching motility assay was carried out as described previously (Park et al., 2014b). Ten microliters of bacterial suspension (1.0 × 108 CFU/ml) was dropped into the center of a PSA plate, dried, and incubated at 28°C for 3 days. The marginal morphology of each dropped colony was observed under a light microscope. This experiment was repeated at least three times with four biological replicates. The margins of the colonies from the wildtype strain were significantly irregular, but XooΔraxST colonies exhibited smooth edges, indicating that RaxST is essential to bacterial twitching motility. In the complementation experiment, XooΔraxST (RaxST) was restored toward to the phenotype of the wildtype strain. Similar to XooΔraxST, the twitching motility of the XooΔomp1X strain was significantly impaired (Park et al., 2014b), indicating that sulfation by RaxST is indispensable for normal twitching motility in Xoo. Flagella-dependent motility was not affected by RaxST (data not shown), although the expression of flagellin (PXO_06165) in the XooΔraxST strain was higher than in the wildtype strain (Table 3).
It is clear that RaxST is critical for full twitching motility in Xoo (Fig. 2). However, because many chemotaxis-related proteins are highly expressed in XooΔraxST, it remains to be investigated whether the impaired twitching motility was affected by the increased proteins directly, indirectly, or both. In addition, XooΔraxST may retain functional pili because PXO_0164 and PXO_02353, which are related to pili/fimbriae biosynthesis, are still expressed in the XooΔraxST strain (Table 3). Therefore, one possible explanation is that uncontrolled high expression of chemotaxis-related proteins in XooΔraxST may lead to abnormal twitching motility. In support of our hypothesis, overexpression of ChpA, a regulatory protein for chemosensory systems in Pseudomonas aeruginosa, has been shown to hamper the normal twitching motility of the wildtype strain (Whitchurch et al., 2004). To determine how tyrosine sulfation by RaxST affects the expression of proteins involved in bacterial motility, each molecular target of RaxST needs to be studied in depth.
In conclusion, this report elucidates the biological function of a tyrosine sulfotransferase, RaxST, using comparative shotgun proteomic analysis and phenotypic observation. The comparative proteomic analysis in this study found correlations between phenotypic changes and the biological functions of target proteins, indicating that a label-free shotgun proteomic technique is a very useful tool for postulating the biological roles of proteins. Our results also reveal that RaxST, the first biochemically characterized prokaryotic tyrosine sulfotransferase, has diverse functions in bacterial biology, including cell motility and signal transduction. However, the specific molecular mechanisms of tyrosine sulfation by RaxST, and its role in biological processes, remain to be characterized. Because Omp1X may function as a porin-like channel required for bacterial motility (Park et al., 2014b), it is possible that tyrosine sulfation by RaxST on Omp1X plays a role in discriminating or differentiating molecules being transported through the sulfated Omp1X on outer membranes of Xoo. The biological functions of RaxX, another substrate of RaxST and an activator of XA21-mediated immunity (Pruitt et al., 2015), are still not fully understood. Therefore, in addition to Omp1X and RaxX, it has to be carried out to identify other substrates of RaxST and to test phenotypes influenced by this protein in Xoo.
Acknowledgments
This work was supported by grants from the Next-Generation BioGreen 21 Program (No. PJ011033012016) of the Rural Development Administration (RDA), and by the Chung-Ang University Excellent Student Scholarship in 2016.
Fig. 1
Comparison and clusters of orthologous groups (COG) analysis of proteins whose expression is altered by RaxST. (A) Venn diagram showing the number of differently expressed proteins in wildtype (red circle) and XooΔraxST (blue circle) strains for the commonly detected proteins in three biological replicates. Four proteins were found only in the wild type; 18 were highly expressed in the wild type; 15 were only detected in XooΔraxST; and 12 were highly expressed in XooΔraxST. Cv, coefficient of variation. (B) COG analysis of proteins whose expression was affected by RaxST. C, energy production and conversion; D, cell cycle control and mitosis; E, amino acid metabolism and transport; F, nucleotide metabolism and transport; G, carbohydrate metabolism and transport; H, coenzyme metabolism; I, lipid metabolism; J, translation; K, transcription; L, replication and repair; M, cell wall/membrane/ envelope biogenesis; N, cell motility; O, post-translational modification, protein turnover, chaperone functions; P, inorganic ion transport and metabolism; Q, secondary structure; R, general functional prediction only; S, function unknown; T, signal transduction mechanisms; U, intracellular trafficking and secretion; V, defense mechanisms.
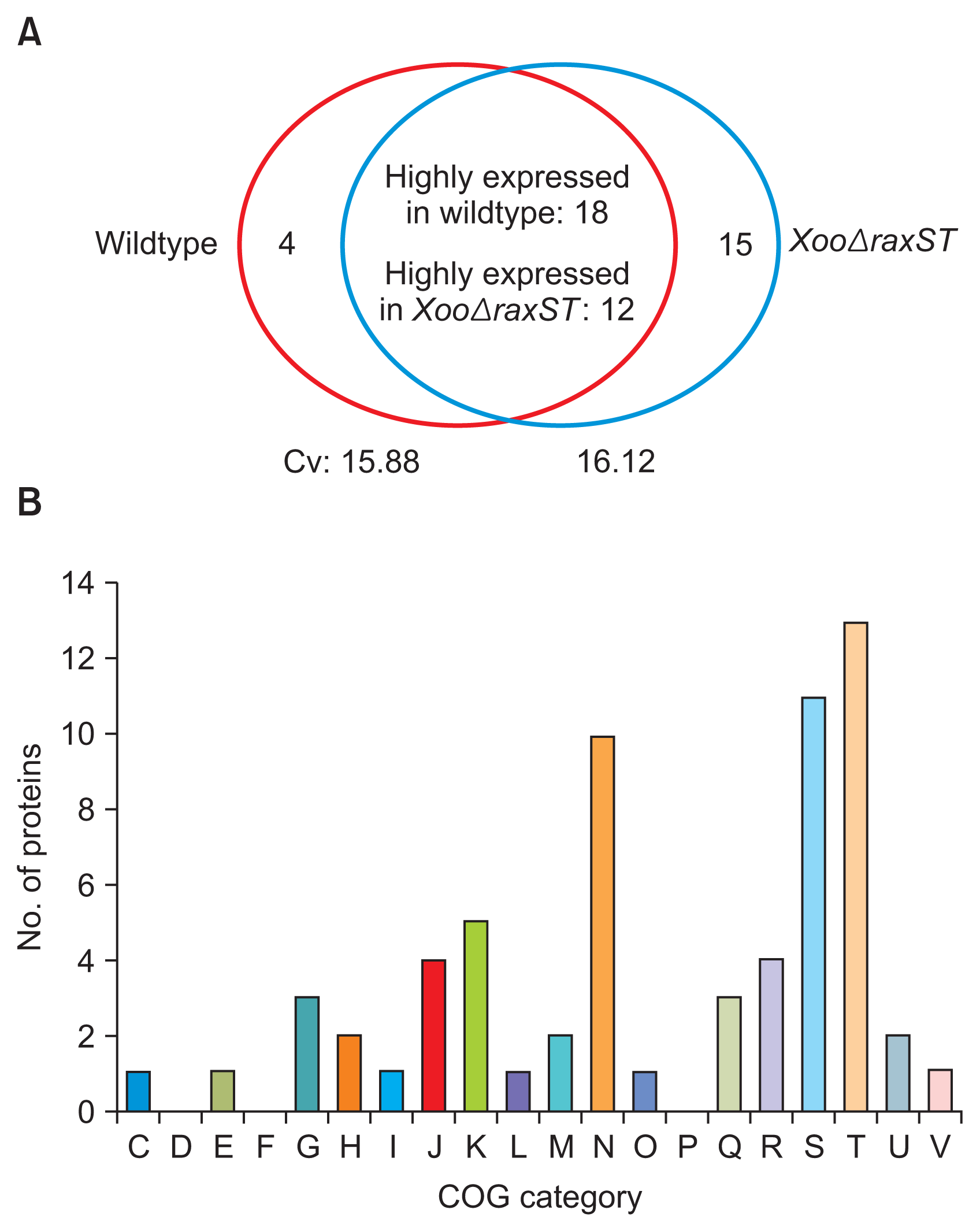
Fig. 2
Twitching motility of the wild type, XooΔraxST, and XooΔraxST (RaxST) strains. Ten microliters of bacterial suspension (1.0 × 108 CFU/ml) was dropped on potato sucrose agar containing 1.5% agar and incubated for 3 days. Twitching motility was evaluated by examination of the marginal shapes under a light microscope. Scale bars = 100 μm.

Table 1
Detected numbers of proteins and PSM in three biological replicates from the label-free shotgun proteomic experiments
| Strain | 1st | 2nd | 3rd | Shared proteins in three biological replicates | |||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| Protein | PSM | Protein | PSM | Protein | PSM | ||
| Wildtype | 753 | 57,274 | 771 | 53,045 | 779 | 60,401 | 661 |
| XooΔ raxST | 768 | 56,642 | 782 | 57,027 | 781 | 60,071 | 662 |
Table 2
Classification of highly expressed (> 1.5 fold) proteins in the wildtype using COG
| COG function | Accession | Locus tag | Gene function | Fold change (ΔraxST/wild) | Included in other group |
|---|---|---|---|---|---|
| C (energy production and conversion) | 188578710 | PXO_03114 | F0F1 ATP synthase subunit epsilon | 1.67 | |
| E (amino acid transport and metabolism) | 188577842 | PXO_01920 | Aminopeptidase N | 1.63 | |
| G (carbohydrate transport and metabolism) | 188579071 | PXO_02746 | Polyvinylalcohol dehydrogenase | 3.24 | |
| 188574985 | PXO_04104 | Beta-glucosidase | 2.37 | ||
| 188579118 | PXO_03298 | Endoglucanase | 1.66 | ||
| H (coenzyme transport and metabolism) | 188578413 | PXO_02519 | 2-amino-3-ketobutyrate coenzyme A ligase | 2.71 | |
| J (translation, ribosomal structure and biogenesis) | 188577053 | PXO_01112 | TrmH family RNA methyltransferase | * | |
| M (cell wall/membrane/envelope biogenesis) | 188575566 | PXO_04698 | OmpA family domain-containing protein | 1.97 | |
| 188577175 | PXO_01117 | UDP-N-acetylglucosamine acyltransferase | 1.86 | ||
| O (post-translational modification, protein turnover, and chaperones) | 188577468 | PXO_01524 | Protein GntY | 1.55 | |
| Q (secondary metabolites biosynthesis, transport, and catabolism) | 188579085 | PXO_03211 | ABC transporter ATP-binding protein | * | |
| 188576165 | PXO_00406 | Acetoacetyl-CoA reductase | 1.61 | I, R | |
| R (general function prediction only) | 188576411 | PXO_00684 | Alkaline phosphatase | 1.55 | |
| S (function unknown) | 188577991 | PXO_01984 | Hypothetical protein | * | |
| 188578598 | PXO_02671 | Adhesin-like protein A | 3.16 | ||
| 188576643 | PXO_00888 | Hypothetical protein | 2.31 | ||
| 188578763 | PXO_03057 | Hypothetical protein | 2.08 | ||
| 188574531 | PXO_03595 | Hypothetical protein | 2.05 | ||
| 188577492 | PXO_01502 | Hypothetical protein | 1.76 | ||
| 188577147 | PXO_01147 | Hypothetical protein | 1.75 | ||
| T (signal transduction mechanisms) | 188578484 | PXO_02599 | Response regulator | 1.58 | |
| V (defense mechanisms) | 188575698 | PXO_04833 | Beta-lactamase | * |
Table 3
Classification of highly expressed (> 1.5 fold) proteins in the XooΔraxST strain using COG analysis
| COG function | Accession | Locus tag | Gene function | Fold change (wild/ΔraxST) | Included in other group |
|---|---|---|---|---|---|
| J (translation, ribosomal structure and biogenesis) | 188577392 | PXO_01601 | Glutathione synthetase | 1.56 | H |
| 188576286 | PXO_00525 | Queuine tRNA-ribosyltransferase | * | ||
| 188578674 | PXO_03147 | ATP-dependent RNA helicase | 1.75 | L, K | |
| K (transcription) | 188578888 | PXO_02929 | Chromosome partitioning protein | 1.63 | |
| 188577000 | PXO_06209 | RNA polymerase sigma factor FliA | * | ||
| N (cell motility) | 188576956 | PXO_06165 | Flagellin | 14.08 | |
| 188577385 | PXO_01608 | Glutamate methyltransferase | 1.53 | T | |
| 188577389 | PXO_01604 | Pilus biogenesis protein | 1.59 | T | |
| 188576008 | PXO_00057 | Chemotaxis-specific methylesterase | * | T | |
| 188576015 | PXO_00050 | Chemotaxis signal transduction protein | * | T | |
| 188576018 | PXO_00047 | Chemotaxis protein | * | T | |
| 188576019 | PXO_00046 | Chemotaxis protein | * | T | |
| 188576031 | PXO_00032 | Chemotaxis protein CheA | * | T | |
| 188577003 | PXO_06212 | Chemotaxis protein CheA | * | T | |
| 188578248 | PXO_02353 | Fimbrial assembly membrane protein | 2.38 | U | |
| Q (secondary metabolites biosynthesis, transport, and catabolism) | 188579087 | PXO_03209 | Toluene tolerance protein | 1.52 | |
| R (general function prediction only) | 188578113 | PXO_02228 | ABC transporter ATP-binding protein | 1.66 | |
| 188577094 | PXO_01199 | GNAT family acetyltransferase | * | ||
| S (function unknown) | 188578099 | PXO_02242 | Hypothetical protein | 1.82 | |
| 188574981 | PXO_04100 | GatB/Yqey | * | ||
| 188576619 | PXO_00915 | Hypothetical protein | * | ||
| 188576905 | PXO_0611 | 1 Hypothetical protein | * | ||
| T (signal transduction mechanisms) | 188576100 | PXO_00466 | Two-component system regulatory protein with GGDEF domain | * | |
| 188577001 | PXO_06210 | Chemotaxis protein CheY | * | ||
| 188576618 | PXO_00916 | Methyl-accepting chemotaxis protein | 8.79 | K | |
| 188576032 | PXO_00031 | Chemotaxis response regulator | * | K | |
| U (intracellular trafficking, secretion, and vesicular transport) | 188579256 | PXO_03487 | Inner membrane protein translocase component YidC | 1.57 |
References
Choi, H, Fermin, D and Nesvizhskii, AI 2008. Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics. 7:2373-2385.



da Silva, FG, Shen, Y, Dardick, C, Burdman, S, Yadav, RC, de Leon, AL and Ronald, PC 2004. Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Mol Plant-Microbe Interact. 17:593-601.


Farzan, M, Mirzabekov, T, Kolchinsky, P, Wyatt, R, Cayabyab, M, Gerard, NP, Gerard, C, Sodroski, J and Choe, H 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 96:667-676.


Gnanamanickam, SS, Priyadarisini, VB, Narayanan, NN, Vasudevan, P and Kavitha, S 1999. An overview of bacterial blight disease of rice and strategies for its management. Curr Sci India. 77:1435-1444.
Han, SW, Lee, SW, Bahar, O, Schwessinger, B, Robinson, MR, Shaw, JB, Madsen, JA, Brodbelt, JS and Ronald, PC 2012. Tyrosine sulfation in a Gram-negative bacterium. Nat Commun. 3:1153



Han, SW, Park, CJ, Lee, SW and Ronald, PC 2008. An efficient method for visualization and growth of fluorescent Xanthomonas oryzae pv. oryzae in planta. BMC Microbiol. 8:164




Hopkins, CM, White, FF, Choi, SH, Guo, A and Leach, JE 1992. Identification of a family of avirulence genes from Xanthomonas oryzae pv. oryzae. Mol Plant-Microbe Interact. 5:451-459.


Kearns, DB, Robinson, J and Shimkets, LJ 2001. Pseudomonas aeruginosa exhibits directed twitching motility up phosphatidylethanolamine gradients. J Bacteriol. 183:763-767.




Kehoe, JW and Bertozzi, CR 2000. Tyrosine sulfation: a modulator of extracellular protein-protein interactions. Chem Biol. 7:R57-R61.


Kovach, ME, Elzer, PH, Hill, DS, Robertson, GT, Farris, MA, Roop, RM 2nd and Peterson, KM 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 166:175-176.


Matsubayashi, Y, Ogawa, M, Morita, A and Sakagami, Y 2002. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science. 296:1470-1472.


Mew, TW 1987. Current status and future prospects of research on bacterial blight of rice. Annu Rev Phytopathol. 25:359-382.

Miller, RM, Tomaras, AP, Barker, AP, Voelker, DR, Chan, ED, Vasil, AI and Vasil, ML 2008. Pseudomonas aeruginosa twitching motility-mediated chemotaxis towards phospholipids and fatty acids: specificity and metabolic requirements. J Bacteriol. 190:4038-4049.




Mitrophanov, AY and Groisman, EA 2008. Signal integration in bacterial two-component regulatory systems. Genes Dev. 22:2601-2611.



Monigatti, F, Hekking, B and Steen, H 2006. Protein sulfation analysis: a primer. Biochim Biophys Acta. 1764:1904-1913.


Park, HJ, Jung, HW and Han, SW 2014a. Functional and proteomic analyses reveal that wxcB is involved in virulence, motility, detergent tolerance, and biofilm formation in Xanthomonas campestris pv. vesicatoria. Biochem Biophys Res Commun. 452:389-394.


Park, HJ, Lee, SW and Han, SW 2014b. Proteomic and functional analyses of a novel porin-like protein in Xanthomonas oryzae pv. oryzae. J Microbiol. 52:1030-1035.



Pruitt, RN, Schwessinger, B, Joe, A, Thomas, N, Liu, F, Albert, M, Robinson, MR, Chan, LJ, Luu, DD, Chen, H, Bahar, O, Daudi, A, De Vleesschauwer, D, Caddell, D, Zhang, W, Zhao, X, Li, X, Heazlewood, JL, Ruan, D, Majumder, D, Chern, M, Kalbacher, H, Midha, S, Patil, PB, Sonti, RV, Petzold, CJ, Liu, CC, Brodbelt, JS, Felix, G and Ronald, PC 2015. The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci Adv. 1:e1500245



Salzberg, SL, Sommer, DD, Schatz, MC, Phillippy, AM, Rabinowicz, PD, Tsuge, S, Furutani, A, Ochiai, H, Delcher, AL, Kelley, D, Madupu, R, Puiu, D, Radune, D, Shumway, M, Trapnell, C, Aparna, G, Jha, G, Pandey, A, Patil, PB, Ishihara, H, Meyer, DF, Szurek, B, Verdier, V, Koebnik, R, Dow, JM, Ryan, RP, Hirata, H, Tsuyumu, S, Won Lee, S, Seo, YS, Sriariyanum, M, Ronald, PC, Sonti, RV, Van Sluys, MA, Leach, JE, White, FF and Bogdanove, AJ 2008. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics. 9:204



Stone, MJ, Chuang, S, Hou, X, Shoham, M and Zhu, JZ 2009. Tyrosine sulfation: an increasingly recognised posttranslational modification of secreted proteins. N Biotechnol. 25:299-317.


Tatusov, RL, Galperin, MY, Natale, DA and Koonin, EV 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36.



Whitchurch, CB, Leech, AJ, Young, MD, Kennedy, D, Sargent, JL, Bertrand, JJ, Semmler, AB, Mellick, AS, Martin, PR, Alm, RA, Hobbs, M, Beatson, SA, Huang, B, Nguyen, L, Commolli, JC, Engel, JN, Darzins, A and Mattick, JS 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol. 52:873-893.





 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




