 |
 |
| Plant Pathol J > Volume 32(5); 2016 > Article |
Abstract
Fusarium graminearum species complex (FGSC) causes Fusarium head blight in small grain cereals. To date, four species (F. graminearum, F. asiaticum, F. boothii, and F. meridionale ) belonging to FGSC frequently occur in Korean cereals. In addition, we first reported the occurrence of additional species (F. vorosii ) within FGSC, which was isolated from barley, corn, and rice in Korea. Phylogenetic analysis of the Fusarium isolates of this group using combined multi-gene sequences confirmed species identification. Moreover, the macroconidia produced by these isolates were morphologically similar to those of the F. vorosii holotype. Chemical analysis indicated that the F. vorosii isolates produced various trichothecenes such as nivalenol and deoxynivalenol with their acetyl derivatives along with zearalenone. Pathogenicity tests demonstrated that all of the F. vorosii isolates examined were pathogenic on barley, corn, and rice with variation in aggressiveness. This study is the first report of F. vorosii in Korean cereals, their pathogenicity towards barley and corn, and their ability to produce trichothecenes and zearalenone.
Fusarium head blight (FHB) is a frequently occurring disease of small grain cereals worldwide. Outbreaks of FHB are usually accompanied by economic loss of cereals and contamination of grains with mycotoxin. Mycotoxins frequently found in cereals are trichothecenes and zearalenone (ZEA), which cause toxic effects in humans and animals (Starkey et al., 2007; Yli-Mattila et al., 2009).
The main causal pathogens of FHB are Fusarium graminearum species complex (FGSC) and F. culmorum (Goswami and Kistler, 2004; Miedaner et al., 2008; Parry et al., 1995). FGSC consists of at least 15 phylogenetically distinct species (i.e., F. graminearum, F. asiaticum, F. acacia-mearnsii, F. aethiopicum, F. boothii, F. mesoamericanum, F. austroamericanum, F. cortaderiae, F. brasilicum, F. meridionale, and five more recently designated species belonging to the Asian clade including F. vorosii, F. gerlachii [Starkey et al., 2007], F. ussurianum [Yli-Mattila et al., 2009], F. louisianense, and F. nepalense [Sarver et al., 2011]). However, F. graminearum sensu strict (hereafter F. graminearum) is a representative species of FGSC. F. graminearum is predominant in wheat and corn mostly in America and Europe (van der Lee et al., 2015; Yli-Mattila, 2010). In Asian countries such as China, Japan, and Korea, F. asiaticum takes the place of F. graminearum because the major crops grown in these areas are rice and barley (Karugia et al., 2009; Lee et al., 2010; Qui et al., 2014; van der Lee et al., 2015; Zhang et al., 2010).
In Korea, only four species of FGSC reportedly occur in cereals (i.e., F. graminearum, F. asiaticum, F. boothii, and F. meridionale [Aoki et al., 2012; Lee et al., 2010]). The prevalence of F. asiaticum was speculated to be due to its association with rice cultivation in Korea (Lee et al., 2009). Upon recent monitoring of Korean barley, corn and rice for the occurrence and diversity of fungi, a new group of Fusarium isolates was detected, which occupied a distinct phylogenetic branch from these previously known species. Upon molecular and mycological characterization, we were able to identify them F. vorosii. The first published report on F. vorosii described that all of the first three F. vorosii isolates originated from wheat heads (two from Japan and one from Hungary) (Starkey et al., 2007). Since then, there have been no reports of F. vorosii isolated from any crop. Here, we report the occurrence of F. vorosii in barley, corn and rice for the first time in Korea, along with a discussion of their toxicity and pathogenicity on crops.
Fusarium species were collected from grain samples as previously described (Lee et al., 2010). The samples were collected from barley, corn, and rice fields throughout Korea during harvest season. One hundred random grains were surface-sterilized by soaking in sodium hypoclorite solution (NaOCl, 1%) for 2 min and washed twice with sterile distilled water, and then placed on potato dextrose agar plates (PDA; Difco Laboratories, Detroit, MI, USA). The plates were incubated at 25°C for 5 days, and the fungal colonies were sub-cultured onto fresh PDA. Each Fusarium isolate was purified through single-spore isolation. For this, the Fusarium strains were inoculated onto carnation leaf agar medium (Leslie and Summerell, 2006) and incubated for 3 weeks under a 12-h fluorescent light/12-h dark cycle to stimulate conidiation. Several isolates of F. graminearum and F. asiaticum used in this study were revived from our laboratory culture collection. Fungal isolates were maintained in 25% glycerol stock cultures at −80°C. Two representative F. vorosii isolates were deposited in Korean Agricultural Culture Collection (KACC; Rural Development Administration, Jeonju, Korea) as F. vorosii KACC 48005 (GWS) and F. vorosii KACC 48006 (#195). Conidial dimension was measured with imaging software (NIS-Elements BR3.0; Nikon Instruments Inc., Tokyo, Japan). For genomic DNA extraction, the Fusarium isolates were grown on PDA plates at 25°C for 5 days, and then the mycelial mass was harvested by scraping. Fungal genomic DNA was extracted as previously described (Lee et al., 2015). For trichothecene analysis, about 10 agar blocks (2 × 2 mm each) from PDA cultures of each isolate were inoculated into 30 g autoclaved rice in 250 ml Erlenmeyer flasks and incubated for 3 weeks.
For species identification of Fusarium isolates and phylogenetic analysis with other Fusarium strains, partial nucleotide sequences of the genes encoding translation elongation factor 1-α (TEF1, 595 bp), TRI101 (858 bp), and MAT1-1-3 (640 bp) were amplified from each isolate as described previously (O’Donnell et al., 2000). For determination of trichothecene chemotype such as deoxynivalenol (DON), nivalenol (NIV), and acetyl derivatives of DON (3-acetyl DON and 15-acetyl DON), the TRI12 and TRI13 genes were amplified as previously described (Lee et al., 2001; Suga et al., 2008). Each PCR tube contained 50 ng of template DNA, 1 × PCR buffer, dNTPs at 0.2 mM each, primers at 10 μM, and 1.25 U ExTaq polymerase (Takara Biomedicals, Shiga, Japan) in a 20 μl reaction volume. PCR products of the DNA marker genes for phylogenetic analysis were purified and sequenced (Macrogen Inc., Seoul, Korea). For phylogenetic analysis, the nucleotide sequences of each marker gene from the Korean isolates were combined and aligned with those of the reference strains retrieved from GenBank (O’Donnell et al., 2000) using Clustal W (Thompson et al., 1994). A total of 2,093 nucleotides from 3 genes were used in analyses by the unweighted pair group method with arithmetic mean (UPGMA) using MEGA ver. 5.2 (Tamura et al., 2011). The robustness of the trees was determined using the full heuristic search option for 1,000 bootstrap replicates. The DNA sequences of newly obtained from Korean F. vorosii isolate (RPB2B, RNA polymerase II second largest subunit) genes were deposited in GenBank (http://www.ncbi.nlm.nih.gov/) under the following accession numbers (KT070885-888 for 11RhGn137-R1, 11RhGb176-R1, 11BhJb195-R1, and 11BhJn3512-R1, respectively).
Quantitative determination of trichothecenes produced on rice substrates was performed by ultra performance liquid chromatography (UPLC) (DON, NIV, and ZEA) or liquid chromatography-mass spectrometry (LC-MS) (acetyl derivatives) analysis. A homogenized grain sample (5 g) was extracted with 20 ml of distilled water (DON and NIV) or with acetonitrile:water (90:10, plus 0.5 g sodium chloride for ZEA). After filtering through a Whatman paper (No. 1), the filtered samples were loaded onto the appropriate IAC column (DONtest for DON and NIV, Zearalatest for ZEA, Vicam) and then eluted with methanol. The eluate was dried under N2 gas and dissolved with water:acetonitrile:methanol (90:5:5 for DON and NIV, or 43:35:22 for ZEA) to be injected into UPLC (Acquity H-class; Waters Corp., Milford, MA, USA). A fluorescence detector and PDA detector were used for ZEA and DON/NIV, respectively. The XSELECT CSH C18 column (2.1 × 100 mm, 3.5 μm particle size) was used for both analyses. For acetyl derivatives analyses, a sample (4 g) was extracted with 17.5 ml water:acetonitrile (7.5:10, plus 0.1% formic acid). The final dried extract was dissolved with 1 ml acetonitrile:water (5:5) before syringe filtration (pore size, 0.2 μm diameter) then loaded onto LC-MS (e2695 separation module and 2489 ultraviolet/visible detector, 3100 mass detector; Waters Corp.). An Agilent zobax SB-Aq C18 (2) column (3.0 × 150 mm, 5 μm particle size) was used with a mobile phase consisting of water:methanol gradient (5 mM ammonium formate added). The MS system was run in ESI+ mode for 15-ADON and ESI- for 3-ADON.
To test the pathogenicity on barley (cv. Youngyang) and rice (cv. Honong) under greenhouse conditions, the central spikelet of each head was spray-inoculated during mid-anthesis with a suspension of the fungal conidia at 1 × 105/ml, obtained from a 5-day-old carboxymethyl cellulose medium culture as previously described (Brown et al., 2014; Han et al., 2007). Five heads per isolate were tested. The sprayed heads were covered with a plastic bag to retain moisture, and then the whole plants were placed in a dew chamber for 3 days. Fourteen days after inoculation, the inoculated heads were harvested for disease scoring. To test ear and stalk rot on corn (cv. Kwangpyeongok), plants grown in pots (stage R1-R2 with silk) were used. Each ear was inoculated by injecting 10 μl of the suspension of fungal conidia (at 1 × 105/ml) using a micropipette tip onto the middle of corn ear through husks into the kernel and stalk by injection into the second internode as described for the ear. Three plants were used per isolate. Three weeks after inoculation, the corn stalks were cut longitudinally to identify stalk discoloration and measure lesion length and husks of the inoculated ears were removed to observe the kernels. Pathogenicity was compared using Tukey multiple comparison method with WINKS SDA 7 (http://www.texasoft.com).
As previously reported (Starkey et al., 2007), colonies of the Korean F. vorosii isolates exhibited characteristics that were indistinguishable from other FGSC species such as F. graminearum and F. asiaticum on PDA (Fig. 1A). Conidial morphology of these isolates generally fit with the original descriptions for width, longitudinal axis, apical beak, symmetry of upper and lower halves, and widest region of 5-septate conidia, but not length (Fig. 1B). Conidial length of Korean F. vorosii was 47.1 μm in mean value (Table 1) while that of the holotype was reported as > 50 μm (Aoki et al., 2012; Starkey et al., 2007). These results suggest that F. vorosii could be morphologically diverse. However, the published description of F. vorosii morphology was only based on three isolates, requiring acquisition of more isolates for confirmation. In addition, it should be stressed that the morphological characteristics cannot be used as the sole source of information to identify species.
Phylogenetic analyses using combined gene sequences revealed a distinct clade of six Fusarium isolates that grouped with the reference stains of F. vorosii. The phylogenetic tree constructed by UPGMA revealed a topology that clearly separated F. vorosii from closely related species, such as F. asiaticum and other members of the FGSC (Fig. 2). Separation of each species of FGSC was strongly supported (bootstrap support value [BS] > 76%). For the F. vorosii isolates, there were three subclades within the F. vorosii clade (BS > 43%) which formed a strongly supported (BS = 80%) monophyletic sister group to F. asiaticum. Close relationship between F. asiaticum and F. vorosii was previously reported (Starkey et al., 2007; Yli-Mattila et al., 2009). These observations suggested that the Korean F. vorosii isolates may be evolutionally related to F. asiaticum.
Chemical analysis demonstrated the ability of all of the isolates to produce NIV with 4-acetyl NIV or DON with 15-ADON, and ZEA on a rice substrate. This is the first report of the production of NIV along with ZEA by F. vorosii. The amounts of NIV produced by five isolates ranged from 2.0 to 6.0 mg/kg, which were comparable to the value of 5.0 mg/kg NIV produced by a F. asiaticum strain (Table 2). These isolates produced greater amounts of 4-acetyl NIV (2.0-135.0 mg/kg) than NIV. However, a Korean isolate of F. asiaticum from barley was previously reported to produce NIV up to 416 μg/g on a rice substrate (Kim et al., 1993). Therefore, the levels of NIV production by these F. vorosii isolates appeared to be much lower on the rice substrate. In the case of DON, the amount produced was also much less than previously reported levels, as a single isolate (#195) produced only 5 mg/kg of DON and 3.5 mg/kg of 15-ADON compared to the mean content of published F. vorosii strains, such as NRRL 37605 (308 and 83.5 ppm for DON and 15-ADON, respectively) (Table 2; Starkey et al., 2007). However, this direct comparison could be misleading as our study used rice cultures for toxin production, whereas the previous study used wheat heads. Therefore, it is appropriate to compare the DON/NIV production capability of each isolate rather than toxin concentration. It is notable that ZEA production was detected in all of the isolates tested (14-1,737 μg/kg). This result extends the toxic diversity of F. vorosii from DON and 15-ADON to NIV and 4-ANIV to ZEA. It is speculated that the dominance of NIV chemotypes among the F. vorosii isolates (5 out of 6) was probably due to cultivation of barley and rice in double-cropping systems in Korea. This cropping system could result in close association with NIV-producing F. asiaticum, which predominates in barley and rice (Kim et al., 1993; Lee et al., 2009).
All six F. vorosii isolates examined induced head blight symptom on barley grains, and ear/stalk rot on corn. The level of pathogenicity was different among hosts, but barley (cv. Youngyang) was the most susceptible (Table 2). All of the NIV-producing isolates were able to cause blight similar to the NIV-producing F. asiaticum isolate. We attempted pathogenicity tests on wheat but could not score the disease, as a wheat isolate (inoculum) caused little blight under the conditions used. Corn was also susceptible, but it was ironic that one isolate from corn (GWS) exhibited less pathogenicity towards corn than the isolates from barley and rice (Table 2, Fig. 3). Both the ear and stalk were infected but the stalk appeared more susceptible to lesion development and was thus easier to score pathogenicity on corn. The DON-producing isolate (#195) was more pathogenic on both corn ear and stalk than most of the NIV chemotype isolates. In corn, most of the F. vorosii isolates (4 of 5) exhibited virulence comparable to those of F. graminearum and F. asiaticum isolates tested. In rice, three of four isolates only developed weak blight symptoms. Therefore, under the test conditions, the Korean F. vorosii isolates were strongly pathogenic to barley and corn and their virulence appeared to be at least equivalent to or greater than the currently occurring Korean F. asiaticum or F. graminearum strains.
The occurrence of F. vorosii in barley, corn, and rice may indicate that F. vorosii has already been adapted to these grain cereals in Korea. Although the frequencies of F. vorosii in these crops were very low (< 1% of among the entire Fusarium population), its aggressiveness and toxic potential could contribute to FHB or cause FHB in grain crops. In Korea, NIV-producing F. asiaticum is a major FHB pathogen (Lee et al., 2009). The origin of Korean F. vorosii, however, is unclear and was not investigated in this study. As the species name of F. vorosii was designated for the first time in 2007, it would be difficult to trace the history of its occurrence in Korea. To date, only three F. vorosii isolates (all 15-ADON chemotype) have been reported (Aoki et al., 2012). One of them was originated in Hungary, and the other two were from Japan. Therefore, the occurrence of 15-ADON chemotype F. vorosii in Korea is likely to be attributable to the close geographical association between Korea and Japan, but this needs to be confirmed by comparison of these isolates. It is interesting that six F. vorosii isoates were detected between 2010 and 2011 samples in Korea. These results suggest that F. vorosii is adapted for growth in Asia. It has been reported that certain species of FGSC, such as F. aethiopicum (Ethiopia), F. acacia-mearnsii (South Africa), and F. cortaderiae (New Zealand), are geographically limited (van der Lee et al., 2015). As we continue to monitor the Fusarium population after 2012, the occurrence of F. vorosii in Korea over subsequent years will be revealed.
The appearance of NIV-producing F. vorosii may indicate that the population of FHB becomes more diversified as the number of NIV-producing species increases. The recent addition of NIV-chemotype species, such as F. gerlachii and F. louisianense to FGSC (Sarver et al., 2011) has contributed to diversification. These NIV-chemotype species appear to have more aggressive pathogenicity towards grain crops than before. It is necessary to conduct consecutive monitoring of Fusarium occurrence in small grain cereals followed by intensive population analyses to determine the global Fusarium population trends. The genetic diversity of F. vorosii isolates in Korea, and their potential for the occurrence of FHB, must also be studied continuously with careful monitoring to detect other species belonging to FGSC in Korean cereals.
Acknowledgments
This study was carried out with the support of “Research Program for Agricultural Science & Technology Development (Project No. PJ008635)”, National Institute of Agricultural Sciences, Rural Development Administration, Republic of Korea.
Fig. 1
Mycelial growth on potato dextrose agar medium (A; top and bottom views are shown in top and bottom panels, respectively, for each isolate) and macroconidial morphology of Fusarium vorosii isolates (B). Scale bars = 20 μm.
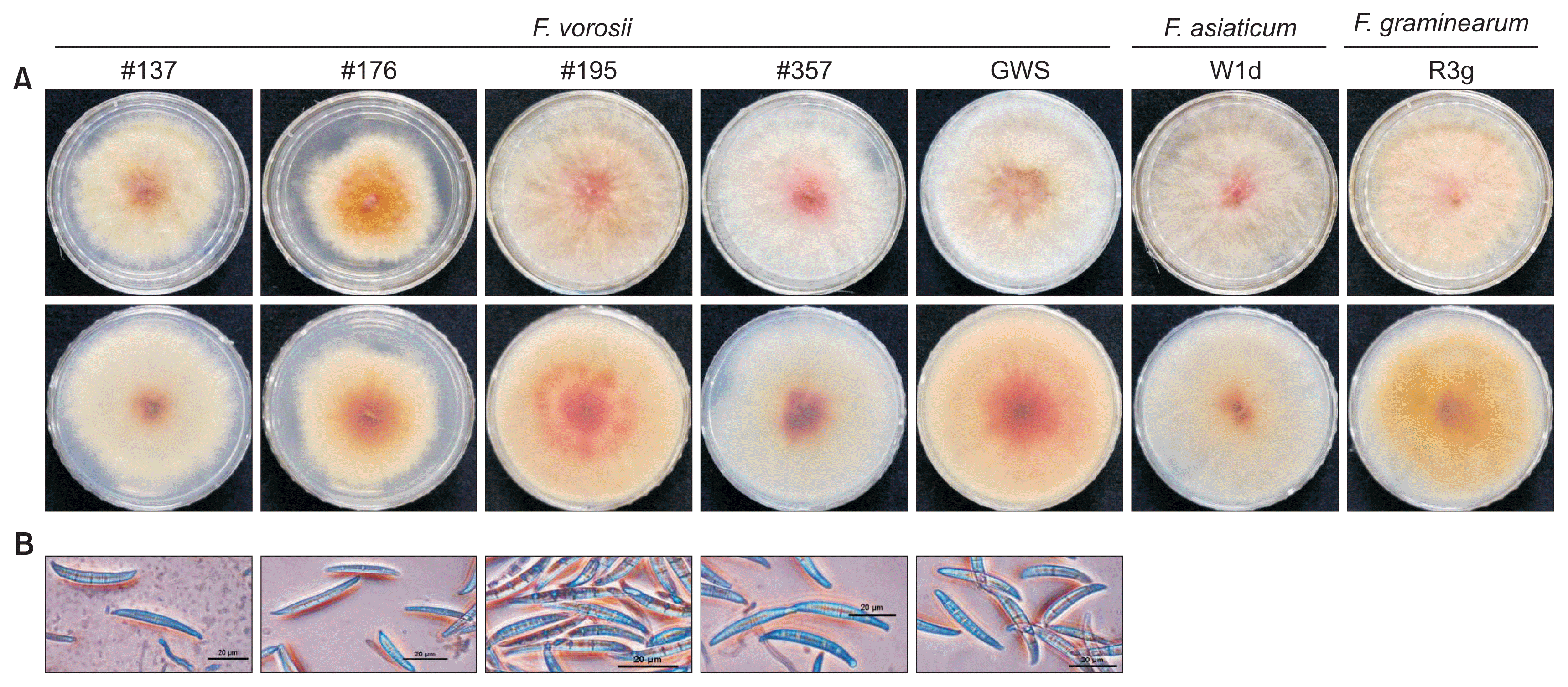
Fig. 2
UPGMA tree inferred from the combined dataset for Fusarium vorosii and its closely related species. The nucleotide sequence of the F. verticillioides 7600 strain was used as the outgroup to root the phylogeny. The bootstrap values are indicated above the internodes and values < 50 are not shown. The numbers of fungal isolates represent our collection number or the NRRL number. The F. vorosii isolates are shown in a gray-shaded box.
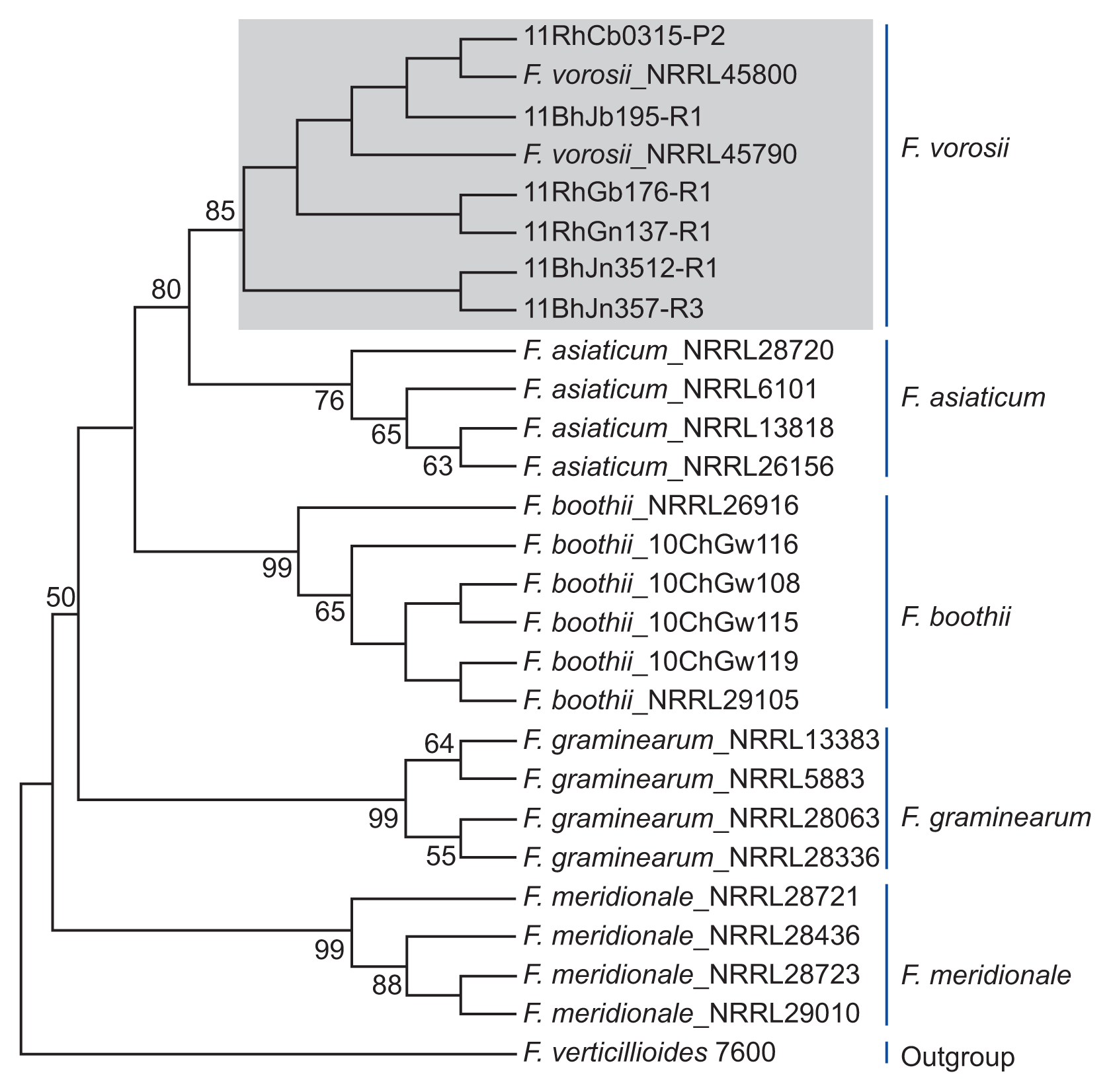
Fig. 3
Pathogenicity of Fusarium vorosii isolates on corn. The left panel shows ears and the right panel shows longitudinal cut of corn stalk. Control received only water. The pictures were taken at 21 days after inoculation.

Table 1
Dimension of macroconidia of Fusarium vorosii isolates
| Isolate | #176 | #195 | #357 | #3512 | #137 | Mean value* |
|---|---|---|---|---|---|---|
| Length (μm) | 47.4 ± 4.1 | 45.2 ± 3.5 | 52.9 ± 6.4 | 49.0 ± 3.6 | 41.2 ± 3.3 | 47.1 ± 4.3 |
| Width (μm) | 5.3 ± 0.5 | 5.2 ± 0.5 | 5.3 ± 0.5 | 5.5 ± 0.6 | 5.3 ± 0.8 | 5.3 ± 0.1 |
Table 2
Pathogenicity and toxin potential of Fusarium vorosii isolates tested in this study
| Isolate | Species | Host | Chemotype | Pathogenicity* (%) | Toxin concentration (mg/kg) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||
| Barley | Corn ear | Corn stem | Rice | NIV | 4-ANIV | DON | 3-ADON | 15-ADON | ZEA† | ||||
| W1d | F. asiaticum | Wheat | 3-ADON | 100a | 55.6abc | 37.2ab | 19.3a | 0 | 0 | 2 | 0.4 | 0 | 24 |
| H5-3 | F. asiaticum | Barley | NIV | 55.4a | 34.4abc | 40.7ab | NT | 5 | 135 | 0 | 0 | 0 | 51 |
| R3g | F. graminearum | Rice | 15-ADON | 97.1a | 37.8abc | 54.8a | 5.4bcd | NT | 0 | 5 | 2.4 | 45 | 1,155 |
| Z-3639 | F. graminearum | Wheat | 15-ADON | 92.9a | 56.1abc | 43.3ab | 13.6ab | NT | 0 | 6 | 0 | 0 | 1,714 |
| GWS | F. vorosii | Corn | NIV | 79.5a | 19.9c | 43ab | 13.0ab | 6 | 10 | NT | 0.1 | 0 | 80 |
| #195 | F. vorosii | Barley | 15-ADON | 92.5a | 74.1a | 60.5a | 10.0abc | NT | 0 | 5 | 0.2 | 3.5 | 171 |
| #357 | F. vorosii | Barley | NIV | 94.5a | 63.5abc | 43.9ab | 0.4cd | 3 | 2 | 0 | 0.1 | 0 | 102 |
| #3512 | F. vorosii | Barley | NIV | 80.5a | 20.7bc | 43.3ab | 7.5bcd | 2 | 94 | NT | 0.1 | 0 | 14 |
| #176 | F. vorosii | Rice | NIV | 64.4a | 69a | 65.9a | NT | 4 | 10 | NT | 0.1 | 0 | 62 |
| #137 | F. vorosii | Rice | NIV | 58.6a | NT | NT | NT | 3 | 124 | 0 | 0.2 | 0.1 | 1,737 |
| Control | - | - | - | 0 | 0 | 12.4b | 0.3d | - | - | - | - | - | - |
NIV, nivalenol; 4-ANIV, 4-acetyl NIV; DON, deoxynivalenol; 3-ADON, 3-acetyl DON; 15-ADON, 15-acetyl DON; ZEA, zearalenone; NT, not tested.
References
Aoki, T, Ward, TJ, Kistler, HC and O’Donnell, KO 2012. Systematics, phylogeny and trichothecene mycotoxin potential of Fusarium head blight cereal pathogens. Mycotoxins. 62:91-102.

Brown, DW, Lee, SH, Kim, LH, Ryu, JG, Lee, S, Seo, Y, Kim, YH, Busman, M, Yun, SH, Proctor, RH and Lee, T 2014. Identification of a 12-gene fusaric acid biosynthetic gene cluster in fusarium species through comparative and functional genomics. Mol Plant-Microbe Interact. 28:319-332.

Goswami, RS and Kistler, HC 2004. Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol. 5:515-525.


Han, YK, Kim, MD, Lee, SH, Yun, SH and Lee, YW 2007. A novel F-box protein involved in sexual development and pathogenesis in Gibberella zeae. Mol Microbiol. 63:768-779.


Karugia, GW, Suga, H, Gale, LR, Nakajima, T, Tomimura, K and Hyakumachi, M 2009. Population structure of the Fusarium graminearum species complex from a single Japanese wheat field sampled in two consecutive years. Plant Dis. 93:170-174.


Kim, JC, Park, AR, Lee, YW, Youn, HJ and Cha, SH 1993. Variation in trichothecene and zearalenone production by Fusarium graminearum isolates from corn and barley in Korea. Korean J Microbiol. 31:312-317.
Lee, J, Chang, IY, Kim, H, Yun, SH, Leslie, JF and Lee, YW 2009. Genetic diversity and fitness of Fusarium graminearum populations from rice in Korea. Appl Environ Microbiol. 75:3289-3295.




Lee, SH, Lee, JK, Nam, YJ, Lee, SH, Ryu, JG and Lee, T 2010. Population structure of Fusarium graminearum from maize and rice in 2009 in Korea. Plant Pathol J. 26:321-327.

Lee, T, Kim, S, Busman, M, Proctor, RH, Ham, H, Lee, S, Hong, SK and Ryu, JG 2015. Rapid detection method for fusaric acid-producing species of Fusarium by PCR. Res Plant Dis. 21:326-329.

Lee, T, Oh, DW, Kim, HS, Lee, J, Kim, YH, Yun, SH and Lee, YW 2001. Identification of deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae by using PCR. Appl Environ Microbiol. 67:2966-2972.




Leslie, JF and Summerell, BA 2006. The fusarium laboratory manual. Wiley-Blackwell, Ames, IA, USA. 388.
Miedaner, T, Cumagun, CJR and Chakraborty, S 2008. Population genetics of three important head blight pathogens Fusarium graminearum, F. pseudograminearum, and F. culmorum. J Phytopathol. 156:129-139.

O’Donnell, K, Kistler, HC, Tacke, BK and Casper, HH 2000. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc Natl Acad Sci U S A. 97:7905-7910.



Parry, DW, Jenkinson, P and McLeod, L 1995. Fusarium ear blight (scab) in small grain-cereals: a review. Plant Pathol. 44:207-238.

Qui, J, Xu, J and Shi, J 2014. Molecular characterization of the Fusarium graminearum species complex in Eastern China. Eur J Plant Pathol. 139:811-823.


Sarver, BA, Ward, TJ, Gale, LR, Broz, K, Kistler, HC, Aoki, T, Nicholson, P, Carter, J and O’Donnell, K 2011. Novel Fusarium head blight pathogens from Nepal and Louisiana revealed by multilocus genealogical concordance. Fungal Genet Biol. 48:1096-1107.


Starkey, DE, Ward, TJ, Aoki, T, Gale, LR, Kistler, HC, Geiser, DM, Suga, H, Toth, B, Varga, J and O’Donnell, K 2007. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet Biol. 44:1191-1204.


Suga, H, Karugia, GW, Ward, T, Gale, LR, Tomimura, K, Nakajima, T, Miyasaka, A, Koizumi, S, Kageyama, K and Hyakumachi, M 2008. Molecular characterization of the Fusarium graminearum species complex in Japan. Phytopathology. 98:159-166.


Tamura, K, Peterson, D, Peterson, N, Stecher, G, Nei, M and Kumar, S 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731-2739.



Thompson, JD, Higgins, DG and Gibson, TJ 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680.



van der Lee, T, Zhang, H, Diepeningen, A and Waalwijk, C 2015. Biogeography of Fusarium graminearum species complex and chemotypes: a review. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 32:453-460.



Yli-Mattila, T 2010. Ecology and evolution of toxigenic Fusarium species in cereals in northern Europe and Asia. J Plant Pathol. 92:7-18.



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




